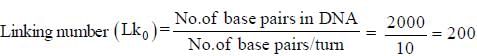Molecular Biology MCQ 2 - Biotechnology Engineering (BT) MCQ
21 Questions MCQ Test Mock Test Series of IIT JAM Biotechnology 2026 - Molecular Biology MCQ 2
Which of the following proteins moves along a dsDNA, unwinds the strands ahead and degrade them, during homologous recombination is
Which of the following codons represent the principle of degeneracy?
The ‘cap’ on a mRNA has a guanine ribonucleotide that is connected in a
Why does enzyme DNA polymerase needs a primer for synthesis of a new strand?
Gene regulation through RNA interference can be described by which of the following statement?
Which of the following are properties of enzyme telomerase ?
Which of the following are needed in the transcription process?
Protein synthesis rates in prokaryotes are limited by the rate of mRNA synthesis. If RNA synthesis occurs at the rate of 50 nucleotides/sec, then rate of protein synthesis occurs at ________ amino acids/sec.
Approximate molecular weight (kDa) of the product after translation of a 390 bases mRNA will be __________KDa
Multiple RNA polymerase transcribes a DNA template, unwinding about 1.5 turns of DNA template per transcription bubble. From the structural information of classical B-DNA the number of transcription bubbles for a 180 base pair DNA molecule will be____________
The value of linking number for a closed circular DNA molecule of 2000 bp, when DNA is underwound by 20 enzymatic turnovers by DNA gysase (+ ATP) _____________ [Answer in Integer]
Assumption: There are 10 base pairs/ turn in relaxed DNA
|
7 docs|34 tests
|