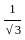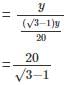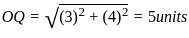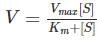CSIR NET Life Sciences Mock Test - 1 - UGC NET MCQ
30 Questions MCQ Test - CSIR NET Life Sciences Mock Test - 1
In a firm the average salary of male employees is Rs. 520 and that of females is Rs. 420. If the overall average salary is Rs. 500, then ratio of male and female employees will be
A dishonest shopkeeper professes to sell pulses at the cost price, but he uses a false weight of 950gm for a kg. Find his gain percent.
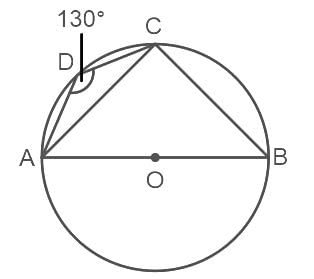
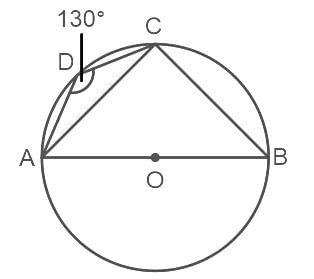
In the given figure, ABCD is a cyclic quadrilateral whose side AB is a diameter of the circle through A, B, C and D. If ∠ADC = 130°, then find ∠CAB.

A man is observing, from the top of a tower, a boat speeding towards the tower from a certain point A, with uniform speed. At that point, angle of depression of the boat with the man's eye is 30° (Ignore man's height). After sailing for 20 seconds, towards the base of the tower (which is at the level of water), the boat has reached a point B, where the angle of depression is 45°. Then the time taken (in seconds) by the boat from B to reach the base of the tower is :
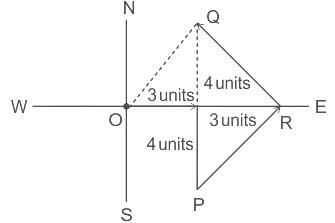
On a planar field, you travelled 3 units East from a point O. Next you travelled 4 units South to arrive at point P. Then you travelled from P in the North-East direction such that you arrive at a point that is 6 units East of point O. Next, you travelled in the North-West direction, so that you arrive at point Q that is 8 units North of point P.
The distance of point Q to point O, in the same units, should be ____
The chart shows forest cover as percentages of total areas of 6 countries over the period 1990-2022 and their land areas (in million km2).
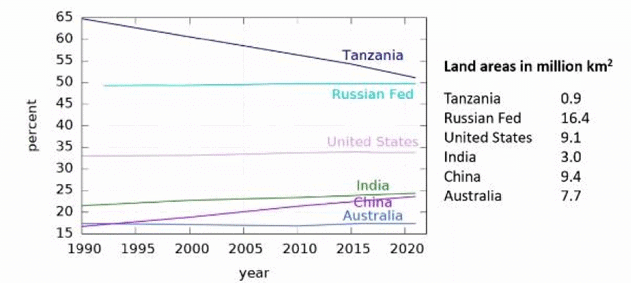
The maximum change in forest area in absolute terms among these countries took place in
Normal human epithelial cells, transformed cells (that originated from stem cells), and epithelial cells transduced with SV40 T antigen (SV40 cells) were cultured for 30 generations. Southern blot analysis was performed using DNA from these cells with radio-labelled probes for histone gene sequences. Which one of the following band patterns would be observed in the autoradiogram?
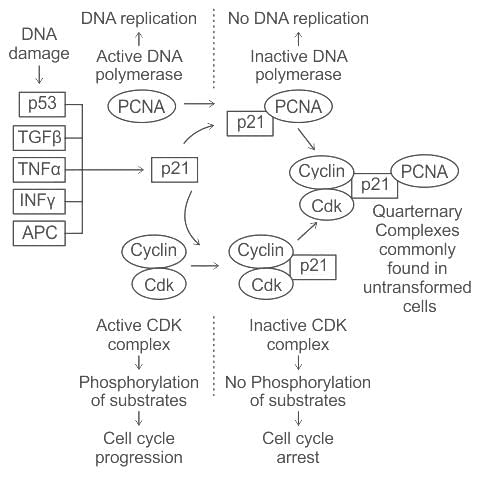
How does p53 contribute to cell cycle regulation when DNA damage is detected?
Which of the following conditions is a peroxisomal disorder?
Two experiments were conducted with an enzyme following Michaelis Menten kinetics at substrate concentrations of 0.5 g/l and 1 g/l. If the enzymatic reaction velocity increases approximately 2 - fold at the higher substrate concentration, the Km for the enzyme would be around:
If a cell has an adequate supply of adenine nucleotides but requires more guanine nucleotides for protein synthesis, which out of these statements are false:
Molecular mass of a protein CANNOT be determined by:
You analyze a cell line deficient in Apaf-1 and observe resistance to apoptosis upon UV radiation. Which of the following proteins is most likely still functioning normally in this cell line?
In 2007, scientists reported the fossil of a deer-like animal in Kashmir, India which is considered the most recent terrestrial ancestor of whales. The name of this fossil is
Secondary metabolites are diverse array of organic compounds in plants. The following are certain statements about secondary metabolites:
A. They protect plants against being eaten by herbivores and against being infected by microbial pathogens.
B. Terpenes, the largest class of secondary metabolites are synthesized by methylerythritol phosphate (MEP) pathway and shikimic acid pathway.
C. The most abundant classes of phenolic compounds in plants are derived from phenylalanine.
D. Alkaloids are nitrogen containing secondary metabolites in plants.
Which one of the following combinations of the above statements is correct?
In an animal experiment;
(i) Electrical stimulation of an area in the brain (A) increased a function (F) which was prevented by systemic injection of adrenergic antagonistic, prazosin.
(ii) Injection of carbachol (cholinergic agonist) into A also increased function F which was, however, not prevented by systemic injection of adrenergic antagonistic, prazosin.
The results are likely to be due to the stimulation of
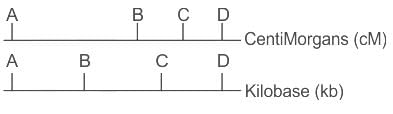
The figure below shows the location of four genes on the genetic map of an organism; the lower panel shows the location of the same group genes on a physical map derived from the nucleotide sequence of the DNA of that organism.What is the reason for the maps to be NOT identical ?
In 'TaqMan' assay for detection of base substitutions (DNA variant), probes (oligonucleotides) with fluorescent dyes at the 5'-end and a quencher at 3'-end are used. While the probe is intact, the proximity of the quencher reduces the fluorescence emitted by reporter dye. If the target sequences (wild type or the variant) are present, the probe anneals to the target sequence, downstream to one of the primers used for amplifying the DNA sequence flanking the position of the variants. For an assay two flanking PCR primers, two probes corresponding to the wild type and variant allele and labelled with two different reporter dyes and quencher were used. During extension the probe may be cleaved by the Taq-polymerase separating the reporter dye and the quencher. Three individuals were genotyped using this assay. Sample for individual I shows maximum fluorescence for the dye attached to the wild type probe, sample for individual II shows maximum fluorescence for the dye attached to variant probe and sample for individual III exhibits equal fluorescence for both the dyes. Which of the following statement is correct?
Certain statements are made concerning nucleic acids and their properties:
A. RNA molecules can exhibit catalytic activity and act as enzymes, known as ribozymes.
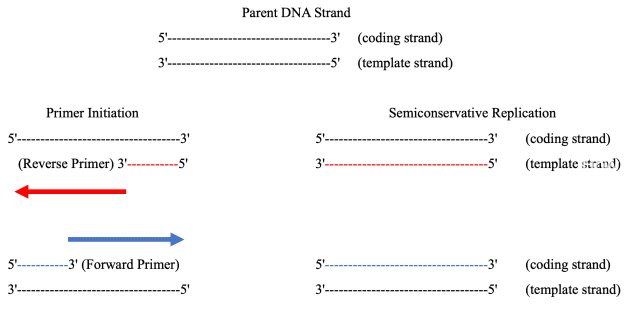
B. All DNA polymerases require a primer to initiate DNA synthesis, which can be either DNA or RNA.
C. A-DNA is the biologically active form that exists under physiological conditions.
D. DNA methylation in eukaryotes occurs predominantly on cytosine bases adjacent to guanine bases, a pattern known as CpG methylation.
E. 5' capping and 3' polyadenylation are modifications specific to eukaryotic tRNA to stabilize it and prevent degradation.
Choose the correct statements.
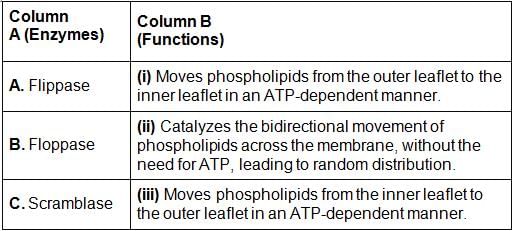
Match the enzymes with their correct functions related to lipid movement across the phospholipid bilayer of cell membranes.
Vegetable seeds purchased from the market germinate when exposed to light with wavelength 660 nm. When followed by a brief exposure to 730 nm they fail to germinate and will germinate when given an exposure of 660 nm. So we can conclude:
Given below is a DNA sequence:
5' – TCGGACTTAGCGTACGATGCTAGTACGGCAT 3'
In the absence of any other affecting parameters such as length, Tm, or GC content, which one of the following pairs of primers would successfully amplify the above DNA fragment?
Choose the option with the combination of all correct statements.
A. Acetylcholine decreases heart rate by acting on muscarinic receptors.
B. Epinephrine increases heart rate by acting on β1-adrenergic receptors.
C. The vagus nerve increases heart rate.
D. Sympathetic stimulation decreases heart rate.
E. Thyroid hormones can increase heart rate.
The characteristic morphological change(s) in cells undergoing apoptosis is/are
A. formation of blebs on cell surface
B. swelling and bursting of cells
C. collapse of the cytoskeleton
D. condensation and fragmentation of nuclear chromatin



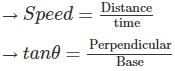
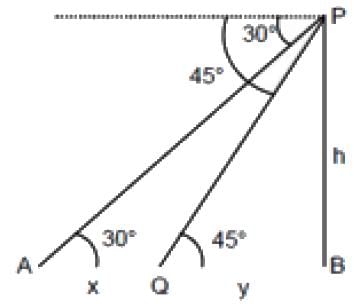
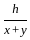 = tan30° =
= tan30° =