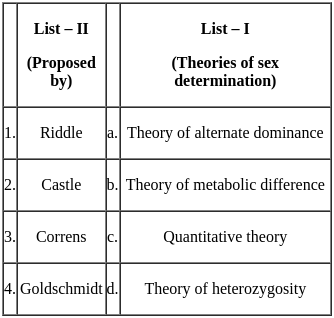
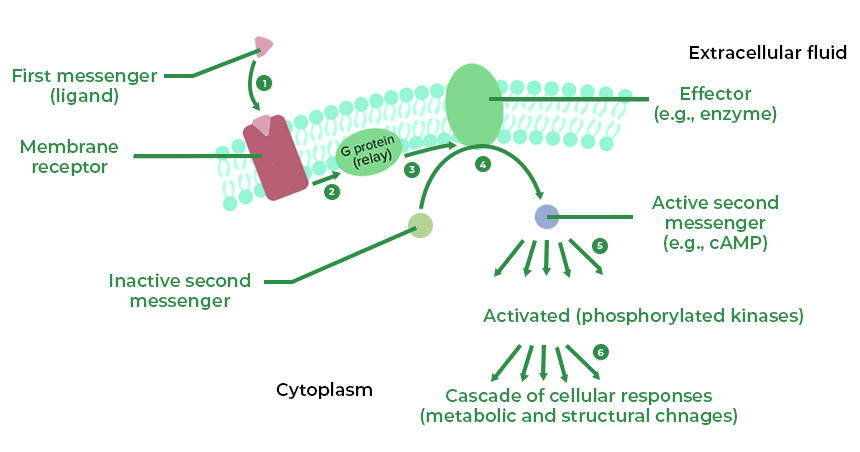
CSIR NET Life Sciences Mock Test - 4 - UGC NET MCQ
30 Questions MCQ Test - CSIR NET Life Sciences Mock Test - 4
A student multiplied a number by 5/7 instead of 7/5. What is the percentage error in the calculation?
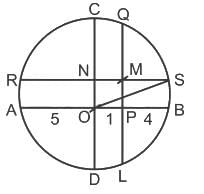
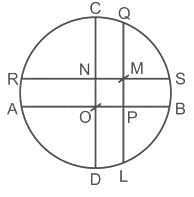
In the given figure, CD and AB are diameters of circle and AB and CD are perpendicular to each other. QL and SR are perpendiculars to AB and CD respectively. Radius of circle is 5 cm, PB ∶ PA = 2 ∶ 3 and CN ∶ ND = 2 : 3. What is the length (in cm) of SM?

The train of a length of 120 m running at a speed of 108 km/hr crosses a platform in 10 sec. Another train running at a speed of 198 km/hr crosses the same platform in 6 seconds. What is the length of the other train?
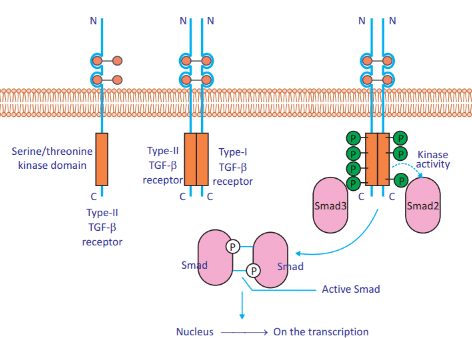
The TGF-β signaling pathway is involved in various cellular processes including cell growth, differentiation, apoptosis, and regulation of the immune system. It begins when TGF-β ligands bind to Type II TGF-β receptors (TβRII), which then recruit and phosphorylate Type I receptors (TβRI). The activated TβRI phosphorylates receptor-regulated SMADs (R-SMADs), which then pair with the co-SMAD (SMAD4). This complex translocates to the nucleus, where it regulates the transcription of target genes." Based on this statement, which of the following conclusions is correct?
Hybridomas are immortalized for production of unlimited supply of monoclonal antibodies by which of the following:
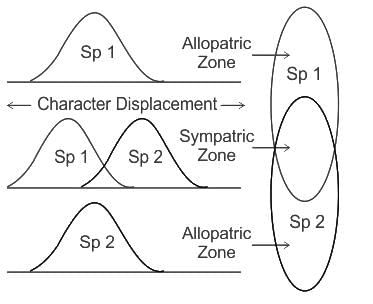
Which of the following statements is the most appropriate example of character displacement?
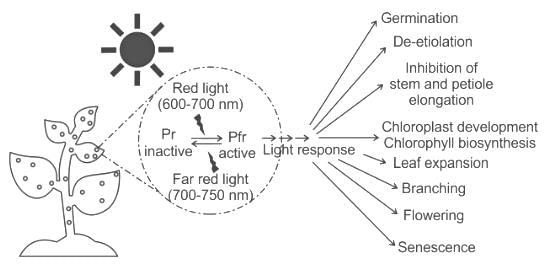
A flash of _____ light during the dark period induces flowering in a long-day plant and the effect is reversed by a flash of _____ light.
On average, how many fragments would a restriction enzyme which recognizes a specific 4 base sequence in DNA be expected to cleave a double-stranded bacteriophage with a genome size of 5,000 bp into?
Choose the correct statement:
Statement 1: IL-2 is critical for the clonal expansion of T cells.
Statement 2: IL-2 supports the growth, survival, and proliferation of activated T cells, including both cytotoxic T cells and helper T cells.
Which of the following is a likely consequence of a loss of function mutation in the gene encoding the enzyme chalcone synthase in flavonoid biosynthesis?
Which one of the following bacteriophages has a genome composed of single stranded circular DNA?
The FISH (Fluorescence In Situ Hybridization) technique is used for which of the following purposes?
Which one of the following does NOT belong to the freshwater ecosystem?
In a three point cross, a recombination event produces the following:
6 Double crossovers
142 Single crossovers
352 Parental genotypes
What would be the percentage recombination between the terminal genes?
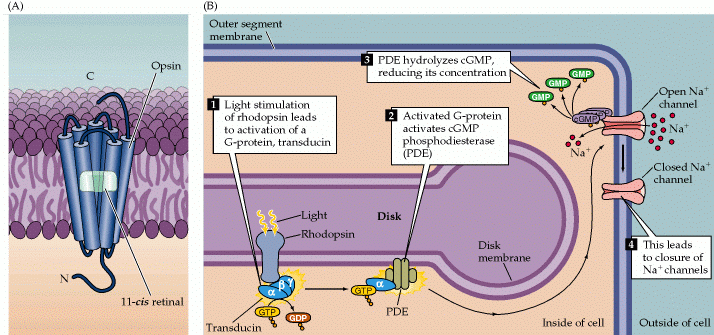
Visual signal transduction cascade is activated by rhodopsin and involves degradation rather than synthesis of which ONE of the following second messenger molecules?
Which one of the following is an insect cell line?
B.F. Skinner conducted experiments where a rat in a box learned to press a lever to receive food. This is an example of:
An enzyme is reported to have a Km of 10 mM and Vmax of 30 mM/s. Assuming Michaelis Menten kinetics, the reaction velocity at a substrate concentration of 20 mM will be:
Which one of the following statements for the lac operon in E. coli is INCORRECT?
Which of these traits is NOT characteristic of r-selected tree species?
You are performing a PCR reaction in which you need to use 20 pmoles of each primer. If both the primers are 20 nucleotides long and the average molecular weight of each nucleotide is 300 Da, the amount of each primer required for 100 µl reaction is:
From the following statements:
A) Coloured images can be obtained by transmission electron microscopy by fluorescent labelling of the specimen.
B) Scanning electron microscopy requires sectioning of the sample.
C) Confocal microscopy uses optical methods to obtain images from a specific focal plane and excludes light from other planes.
D) Differential-interference microscopy relies on interference between polarized light due to differences in the refractive index of the object and surrounding medium.
E) Visualization in epifluorescence microscopy requires staining by heavy metal atoms.
Choose the combination with two correct and one incorrect statement:
Which two of the following vaccines are products of genetic engineering?
i. Diphtheria vaccine
ii. Hepatitis B vaccine
iii. Foot and mouth disease vaccine
iv. Influenza vaccine
v. Tetanus vaccine
The correct combination is :