Culture Media & Methods, Identification of Bacteria by Conventional Chapter Notes | Microbiology - NEET PG PDF Download
| Table of contents |

|
| Culture Media |

|
| Culture Methods |

|
| Identification of Bacteria |

|
| Molecular Techniques for Diagnosis |

|
| Microbial Typing |

|
Culture Media
Concentration of Agar:
- Solid agar is typically used at a concentration of 1-2%, with 2% for Japanese agar and 1.2% for New Zealand agar.
- Semisolid agar contains 0.5% agar.
- To inhibit Proteus swarming, solid agar should be at a concentration of 6%.
Simple/Basal Media
- Peptone Water: Contains 1% peptone, 0.5% NaCl, and water.
- Nutrient Broth: Made by adding 1% meat extract to peptone water.
- Nutrient Agar: Prepared by adding 2% agar to nutrient broth.
Differential Media
- MacConkey Agar
- CLED Agar
Basic Constituents of Culture Media
- Peptone:. blend of partially digested proteins.
- Agar: Used for solidifying culture media; it has no nutritional value.
- Agar is derived from red algae, specifically Gelidium and Gracilaria species.
- Agar is preferred over gelatin because of its bacteriological inertness, with a melting point of about 85-90°C and a solidifying temperature of approximately 32-40°C.
Enriched Media
- Enriched media are used to promote the growth of fastidious bacteria by adding extra nutrients to a basic medium.
- Blood agar is made by adding 5–10% sheep blood to molten nutrient agar at 45°C. It is used to test the hemolytic properties of bacteria.
- Chocolate agar is a heated version of blood agar, made by adding blood to molten nutrient agar at 70°C. It is richer than blood agar and can support the growth of Haemophilus influenzae.
- Loeffler’s serum slope is a selective medium used for isolating Corynebacterium diphtheriae.
- Blood culture media are used for blood cultures and include:
- Monophasic medium: Composed of brain heart infusion (BHI) broth.
- Biphasic medium: Contains a liquid phase (BHI broth) and a solid agar slope (BHI agar).
Enrichment Broth
- Selenite F and Tetrathionate broth: Used for growing Salmonella and Shigella pathogens while inhibiting normal flora.
- Alkaline peptone water (APW): Selective for Vibrio cholerae by allowing its growth and suppressing other organisms.
Selective Media
- Lowenstein Jensen (LJ) medium. Used for isolating Mycobacterium tuberculosis from clinical samples.
- Thiosulphate Citrate Bile Salt Sucrose (TCBS) agar. Selective for Vibrio species, allowing their growth while inhibiting other bacteria.
- DCA (Deoxycholate Citrate Agar) and XLD (Xylose Lysine Deoxycholate) agar. Used for isolating enteric pathogens such as Salmonella and Shigella from stool samples.
- Potassium tellurite agar (PTA): Selective for diphtheroids and other organisms, including Corynebacterium diphtheriae, by allowing their growth on the medium.
- Wilson Blair bismuth sulphite medium. Used for isolating Salmonella Typhi from clinical specimens, particularly stool samples.
Transport Media
Transport media are specifically designed to transport specimens containing fragile organisms or when there might be delays in processing. In these media, bacteria do not grow; they merely remain alive.
Transport media used for common bacteria
- Streptococcus: Pike’s medium
- Neisseria: Amies medium, Stuart’s medium, VR (Venkatraman-Ramakrishnan) medium, Cary Blair medium, and autoclaved sea water
- Shigella and Salmonella: Buffered glycerol saline and Cary Blair medium
These media aid in differentiating between lactose fermenters and non-lactose fermenters using indicators.
MacConkey agar:
- Differentiates organisms into LF (lactose fermenters, producing pink colonies, e.g., E. coli and Klebsiella) and NLF (non-lactose fermenters, producing colourless colonies, e.g., Shigella and Salmonella).
CLED agar (Cysteine Lactose Electrolyte-Deficient agar):
- Similar to MacConkey agar in differentiating between LF and NLF.
- Used as an alternative to blood agar and MacConkey agar for processing urine specimens.
Advantages over MacConkey agar:
- Less inhibitory, supporting gram-positive bacteria (except β-hemolytic Streptococcus) and Candida.
- Prevents swarming of Proteus, an advantage over blood agar.
Anaerobic Culture Media
Anaerobic culture media are specially designed to create an environment with low oxygen levels, which is essential for the growth of certain microorganisms that thrive in oxygen-free conditions, such as Clostridium species. These media reduce the redox potential, making it conducive for anaerobic organisms.
Here are some commonly used types of anaerobic media:
- BHIS Agar: This is Brain Heart Infusion Agar enriched with supplements like vitamin K and hemin to support the growth of anaerobic bacteria.
- Thioglycollate Broth:. liquid medium that contains thioglycollate, which helps to create a low-oxygen environment, suitable for anaerobic bacteria.
- Anaerobic Blood Agar: Blood agar plates that are prepared under anaerobic conditions to support the growth of a wide range of bacteria, including anaerobes.
- Neomycin Blood Agar:. type of blood agar that contains neomycin, an antibiotic that helps to suppress the growth of certain aerobic bacteria, allowing anaerobes to flourish.
- Egg Yolk Agar:. nutrient-rich agar that contains egg yolk, providing essential nutrients for the growth of specific anaerobic bacteria.
- Phenylethyl Agar: This agar contains phenylethyl alcohol, which inhibits the growth of gram-negative bacteria, making it easier to isolate anaerobic organisms.
Robertson’s Cooked Meat (RCM) Broth: This broth contains pieces of cooked meat, usually beef heart, which supply important nutrients like glutathione and unsaturated fatty acids, promoting the growth of anaerobic bacteria.
Culture Methods
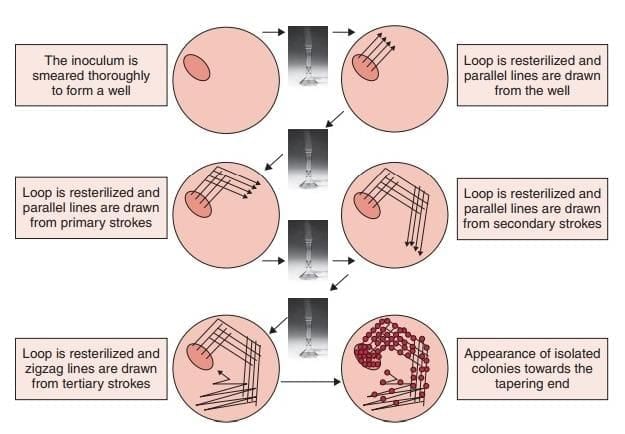
Aerobic Culture Techniques
- Streak Culture: Performed using a loop with intermittent heating; the standard method for routine bacterial isolation.
- Lawn/Carpet Culture: Creates a uniform bacterial layer, ideal for antimicrobial susceptibility testing (disk diffusion), bacteriophage typing, and producing bacterial antigens or vaccines.
- Stroke Culture: Involves zigzag inoculation with a straight wire, used for tests like citrate, urease, and Triple Sugar Iron (TSI).
- Stab Culture: Involves piercing semisolid agar with a straight wire, used for:
- Maintaining stock cultures
- Oxidation-Fermentation (OF) test
- Mannitol motility medium
- Nutrient agar semisolid butts
- TSI (combined with stroke culture)
- Liquid Culture:
- Applications: Blood culture, sterility testing.
- Advantages:
- Effective for low bacterial loads
- Neutralizes antibiotics in specimens (e.g., blood) via dilution
- Yields large bacterial quantities
- Demonstrates bacterial growth curves
- Disadvantages: Cannot isolate pure cultures from mixed inocula.
- Pour-Plate Culture: A quantitative method to estimate viable bacterial counts.
Incubation Conditions
- Most pathogens grow optimally at 37°C.
- Candle Jar: Provides a capnophilic environment (3–5% CO₂), suitable for Brucella abortus, Streptococcus, Pneumococcus, and Gonococcus.
- Microaerophilic Bacteria: Campylobacter and Helicobacter require 5% oxygen.
Anaerobic Culture Techniques
- Vacuum Production: Creates an oxygen-free environment.
- Oxygen Displacement/Combustion:
- McIntosh and Fildes Anaerobic Jar: Manually evacuates air, replaced with hydrogen.
- Anoxomat System: Automated version of the same principle.
- GasPak System: Chemically absorbs oxygen (e.g., using alkaline pyrogallol). Anaerobiosis indicators:
- Chemical: Reduced methylene blue
- Biological: Pseudomonas
- Anaerobic Glove Box: A sealed chamber for anaerobic work.
- Reducing Agents: Include glucose, thioglycollate, meat, cysteine, and ascorbic acid.
- PRAS Media: Prereduced, anaerobically sterilized media.
Microorganism Preservation
Short-Term Methods:
- Sub-culturing
- Storage in glycerol or sterile distilled water
- Freezing at –20°C
- Drying (for molds and spores)
Long-Term Methods:
- Ultra-low temperature freezing
- Lyophilization (freeze-drying)
Identification of Bacteria
- Conventional methods (culture and identification by biochemical reactions)
- Automated culture techniques
- Molecular methods
Automated Culture Techniques
Traditional culture methods often yield poor results because of low bacterial counts. As a solution, various automated blood culture techniques have been developed and used over the past decade.
Advantages of Automated Blood Culture Techniques:
- Continuous Monitoring: Automated systems monitor cultures continuously, checking for growth every 15 to 20 minutes.
- Increased Sensitivity: These techniques are more sensitive, leading to higher yields of bacteria.
- Faster Results: Automated systems provide quicker results and reduce the intensity of manual labor involved.
Disadvantages of Automated Blood Culture Techniques:
- High Cost: Automated systems are expensive to implement and maintain.
- Colony Morphology: The use of liquid medium in automated systems prevents examination of colony morphology.
- Mixed Cultures: Automated systems do not provide separate detection for mixed cultures.
- Safety Concerns: There are potential safety concerns associated with BACTEC systems.
Molecular Techniques for Diagnosis
- Nucleic Acid Amplification Tests (NAATs) are advanced methods used to detect and amplify specific DNA or RNA sequences. These include:
- Polymerase Chain Reaction (PCR) and its variants, such as Real-time PCR, which allows for the simultaneous amplification and quantification of nucleic acids.
- Ligase Chain Reaction (LCR), a method that uses DNA ligase to amplify nucleic acid sequences.
- Transcription-Mediated Amplification (TMA), which amplifies RNA targets by reverse transcription followed by amplification.
Automated Culture Systems in Diagnostic Bacteriology
For Bacterial Culture:
- BACTEC: This system was originally based on radiometric detection but has evolved to use fluorescent-based detection techniques to identify bacterial growth.
- BacT/Alert: This system detects the presence of bacteria by measuring the CO2 released during bacterial metabolism, which causes a change in pH detected by colourimetric methods.
- ESP Culture System: In this system, the CO2 produced by bacteria leads to a pressure change that is detected by manometric methods, indicating bacterial growth.
For Bacterial Identification:
- Phoenix Bacterial Identification System:. system used for the identification of bacteria based on their biochemical characteristics.
- MALDI-TOF (Matrix-Assisted Laser Desorption/Ionization Time-of-Flight) Mass Spectrometry: For example, the VITEK MS system uses MALDI-TOF technology to identify bacteria by analyzing the protein profile of bacterial cells.
- VITEK 2: This system combines bacterial identification and antimicrobial susceptibility testing, providing rapid results for clinical microbiology.
- Microscan Walkaway System: An automated system for the identification and susceptibility testing of bacteria and fungi based on microdilution technology.
For Mycobacterium tuberculosis:
- MGIT (Mycobacterial Growth Indicator Tube):. liquid culture system used for the detection and growth of Mycobacterium tuberculosis and other mycobacteria.
- Nucleic Acid Sequence-Based Amplification (NASBA):. method for the amplification of RNA targets, useful in the detection of Mycobacterium tuberculosis.
- Strand Displacement Amplification (SDA):. nucleic acid amplification technique that can be used for the detection of Mycobacterium tuberculosis.
Polymerase Chain Reaction (PCR)
PCR is a technique used in molecular biology to produce millions of copies of a specific DNA segment from just one or a few copies. It was invented by Kary B. Mullis.
Principle of PCR. PCR involves three main steps:
- DNA Extraction: This is the process of obtaining DNA from an organism.
- Amplification of Extracted DNA: The extracted DNA is amplified through 30–35 cycles in a thermocycler over approximately 3–4 hours. Each cycle consists of three stages:
- Denaturation at 95°C: This step separates the double-stranded DNA (dsDNA) into two single-stranded DNA (ssDNA) strands.
- Primer Annealing at 55°C:. primer, a short piece of DNA matching a specific sequence of the target DNA, attaches to the ssDNA.
- Extension of the Primer at 72°C: This process is carried out by the enzyme Taq Polymerase.
- Gel Electrophoresis: The amplified DNA is separated by size through electrophoresis, creating visible bands under UV light.
Advantages of PCR. PCR offers several benefits over traditional culture methods:
- Increased Sensitivity: PCR can amplify very few copies of specific DNA, making it highly sensitive.
- Enhanced Specificity: PCR uses primers that target specific DNA sequences from the organism.
- Organism Detection: PCR can identify organisms from samples and confirm culture isolates.
- Detection of Fastidious Organisms: PCR can find organisms that are difficult to culture using standard methods.
- Detection of Drug Resistance Genes: PCR can detect genes associated with drug resistance, such as the MecA gene in Staphylococcus aureus.
- Identification of Genetic Diseases: PCR can identify genetic disorders, including sickle cell anaemia and phenylketonuria.
- Conventional PCR: This method is designed for DNA detection. Detecting RNA requires a different process, such as RT-PCR.
- Qualitative Results: PCR provides qualitative results, while quantification is done using real-time PCR.
- Viability Limitations: PCR does not distinguish between live and dead organisms.
- False Positives: Contamination with environmental DNA can lead to false positive results.
- False Negatives: Inhibitors present in samples like blood or faeces can cause false negative results.
Modification of PCR
Reverse transcriptase PCR (RT-PCR):
- This method is used for amplifying RNA.
- After extracting RNA, the first step involves adding the reverse transcriptase enzyme, which converts RNA into DNA.
- The steps for amplifying DNA and documenting it on a gel are the same as those in conventional PCR.
- This technique is beneficial for detecting RNA viruses or 16S rRNA genes from different organisms.
- Nested PCR: The products from the first PCR round undergo a second amplification using a different primer for another gene of the same organism.
PCR consists of three basic steps:
- Multiplex PCR: This technique employs multiple primers to detect several DNA sequences from different organisms in a single reaction.
- Bacteriophage typing is performed for Salmonella Typhi.
- Increased sensitivity: A double amplification round produces a larger quantity of DNA.
- More specific: Two primers targeting the same organisms enhance specificity.
- Disadvantage: Higher contamination risk in PCR tubes can result in false positive outcomes.
- Syndromic approach: This method is used to diagnose infections caused by multiple organisms.
- There is a risk of contamination from environmental DNA in the reaction tubes.
Real-time PCR (rt-PCR)
Real-time PCR, though more costly than conventional PCR, offers several advantages:
- Quantitative: Enables monitoring of treatment response, e.g., for HIV or HBV.
- Faster: Amplification is visualized during the cycle, reducing overall time.
- Low Contamination Risk: Minimizes contamination due to closed-system processing.
- High Sensitivity and Specificity: Provides superior accuracy in detecting target sequences.
Detection Methods for Amplification Products
- Nonspecific Detection: Uses SYBR Green dye, which binds nonspecifically to any nucleic acid.
- Specific Detection: Utilizes fluorescent-labeled DNA probes, including:
- TaqMan (Hydrolysis) Probe
- Molecular Beacon
- FRET (Fluorescence Resonance Energy Transfer) Probe
Microbial Typing
Microbial typing goes beyond identifying organisms by their species; it aims to:
- Investigate outbreaks
- Determine sources and pathways of infections
Genotypic methods are usually more reliable and reproducible than phenotypic ones, though they tend to be more expensive.
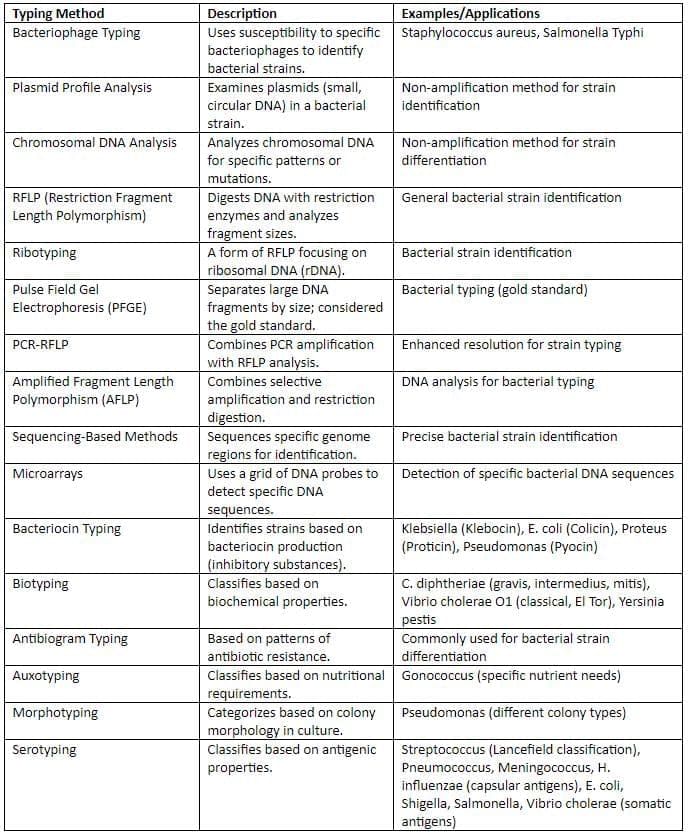
Typing Methods
|
75 docs|5 tests
|
FAQs on Culture Media & Methods, Identification of Bacteria by Conventional Chapter Notes - Microbiology - NEET PG
| 1. What are the different types of culture media used in microbiology? |  |
| 2. What are the conventional methods for identifying bacteria? |  |
| 3. How do molecular methods enhance the identification of bacteria? |  |
| 4. What is microbial typing and why is it important? |  |
| 5. What factors should be considered when selecting culture methods for bacterial identification? |  |




















