Nucleic Acids: Structure & DNA Replication | Biology for Grade 12 PDF Download
The Function of DNA & RNA
- Deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) are both types of nucleic acid
- DNA and RNA are both found in all living cells
- DNA and RNA are both needed to build proteins, which are essential for the proper functioning of cells
- DNA and RNA are both important information-carrying molecules, although their functions are slightly different
DNA
- The function of DNA is to hold or store genetic information
- DNA is the molecule that contains the instructions for the growth and development of all organisms
RNA
- The function of RNA is to transfer the genetic code found in DNA out of the nucleus and carry it to the ribosomes in the cytoplasm
- Ribosomes are where proteins are produced - they ‘read’ the RNA to make polypeptides (proteins) in a process known as translation
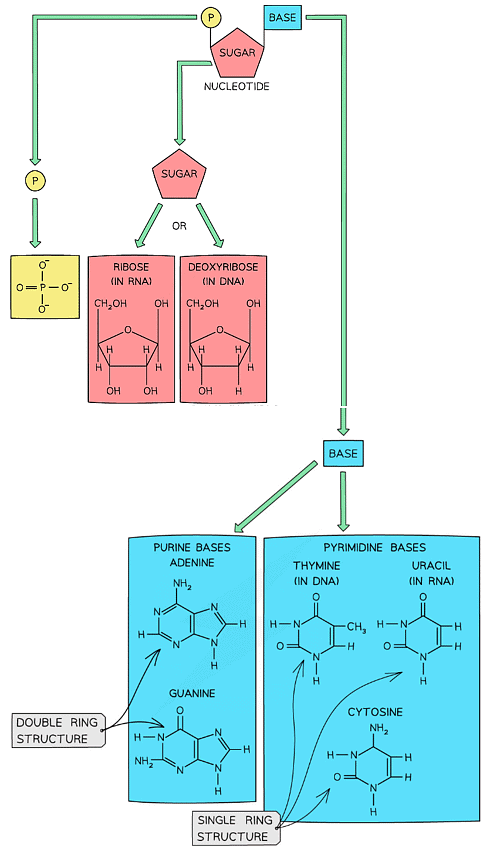
Nucleotide Structure & the Phosphodiester Bond
- Both DNA and RNA are polymers that are made up of many repeating units called nucleotides
- Each nucleotide is formed from:
- A pentose sugar (a sugar with 5 carbon atoms)
- A nitrogen-containing organic base
- A phosphate group

The basic structure of a nucleotide
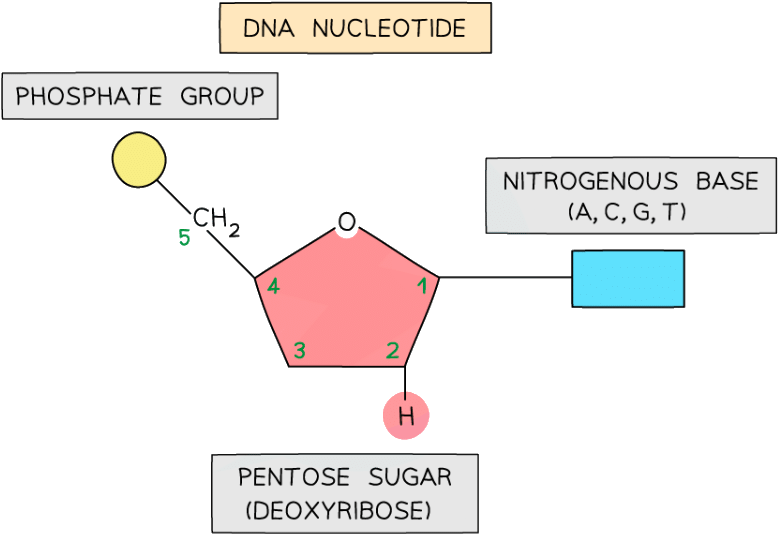
DNA nucleotides
- The components of a DNA nucleotide are:
- A deoxyribose sugar with hydrogen at the 2' position
- A phosphate group
- One of four nitrogenous bases - adenine (A), cytosine(C), guanine(G) or thymine(T)
RNA nucleotides
- The components of an RNA nucleotide are:
- A ribose sugar with a hydroxyl (OH) group at the 2' position
- A phosphate group
- One of four nitrogenous bases - adenine (A), cytosine(C), guanine(G) or uracil (U)
- The presence of the 2' hydroxyl group makes RNA more susceptible to hydrolysis
- This is why DNA is the storage molecule and RNA is the transport molecule with a shorter molecular lifespan
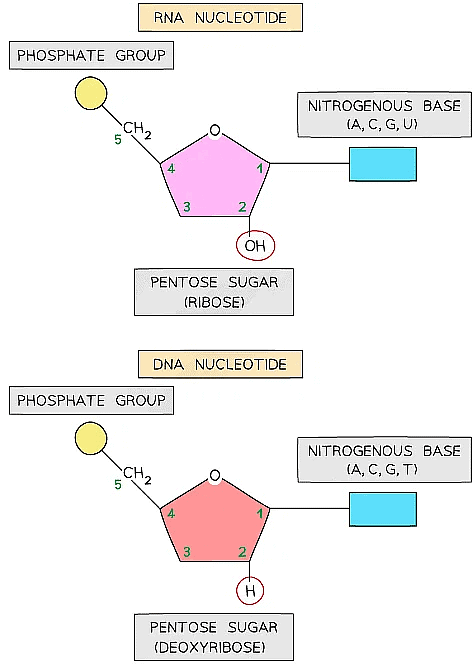 An RNA nucleotide (top) compared with a DNA nucleotide (bottom)
An RNA nucleotide (top) compared with a DNA nucleotide (bottom)
Purines & Pyrimidines
- The nitrogenous base molecules that are found in the nucleotides of DNA (A, T, C, G) and RNA (A, U, C, G) occur in two structural forms: purines and pyrimidines
- The bases adenine and guanine are purines – they have a double ring structure
- The bases cytosine, thymine and uracil are pyrimidines – they have a single ring structure

The molecular structures of purines and pyrimidines are slightly different
Nucleotide Structure Table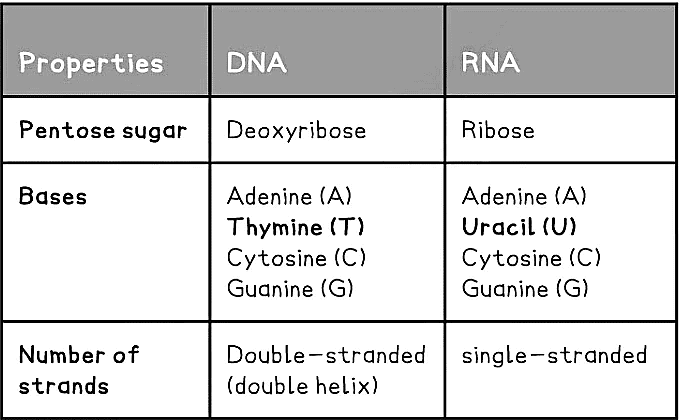 Phosphodiester bonds
Phosphodiester bonds
- DNA and RNA are polymers (polynucleotides), meaning that they are made up of many nucleotides joined together in long chains
- Separate nucleotides are joined via condensation reactions
- These condensation reactions occur between the phosphate group of one nucleotide and the pentose sugar of the next nucleotide
- A condensation reaction between two nucleotides forms a phosphodiester bond
- It is called a phosphodiester bond because it consists of a phosphate group and two ester bonds (phosphate with double bond oxygen attached - oxygen - carbon)
- The chain of alternating phosphate groups and pentose sugars produced as a result of many phosphodiester bonds is known as the sugar-phosphate backbone (of the DNA or RNA molecule)

The Structure of DNA
- The nucleic acid DNA is a polynucleotide – it is made up of many nucleotides bonded together in a long chain

A DNA nucleotide
- DNA molecules are made up of two polynucleotide strands lying side by side, running in opposite directions – the strands are said to be antiparallel
- Each DNA polynucleotide strand is made up of alternating deoxyribose sugars and phosphate groups bonded together to form the sugar-phosphate backbone. These bonds are covalent bonds known as phosphodiester bonds
- The phosphodiester bonds link the 5-carbon of one deoxyribose sugar molecule to the phosphate group from the same nucleotide, which is itself linked by another phosphodiester bond to the 3-carbon of the deoxyribose sugar molecule of the next nucleotide in the strand
- Each DNA polynucleotide strand is said to have a 3’ end and a 5’ end (these numbers relate to which carbon on the pentose sugar could be bonded with another nucleotide)
- As the strands run in opposite directions (they are antiparallel), one is known as the 5’ to 3’ strand and the other is known as the 3’ to 5’ strand
- The nitrogenous bases of each nucleotide project out from the backbone towards the interior of the double-stranded DNA molecule
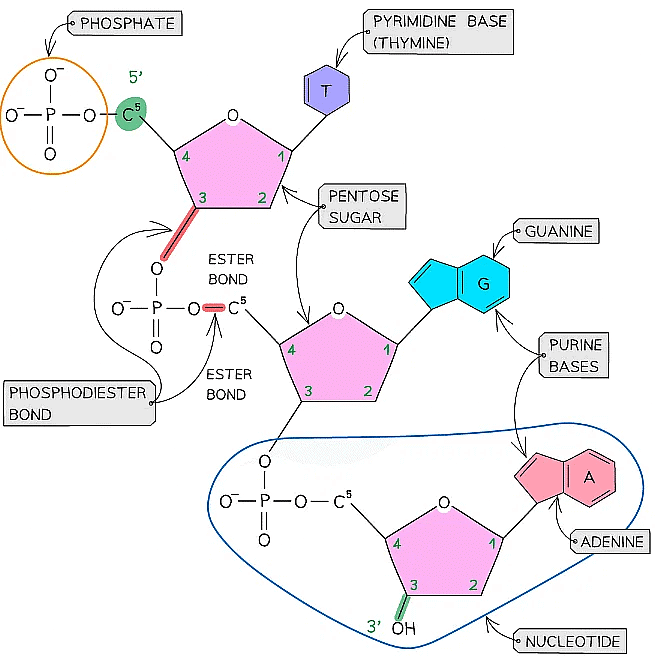 A single DNA polynucleotide strand showing the positioning of the ester bonds
A single DNA polynucleotide strand showing the positioning of the ester bonds
Hydrogen bonding
- The two antiparallel DNA polynucleotide strands that make up the DNA molecule are held together by hydrogen bonds between the nitrogenous bases
- These hydrogen bonds always occur between the same pairs of bases:
- The purine adenine (A) always pairs with the pyrimidine thymine (T) – two hydrogen bonds are formed between these bases
- The purine guanine (G) always pairs with the pyrimidine cytosine (C) – three hydrogen bonds are formed between these bases
- This is known as complementary base pairing
- These pairs are known as DNA base pairs
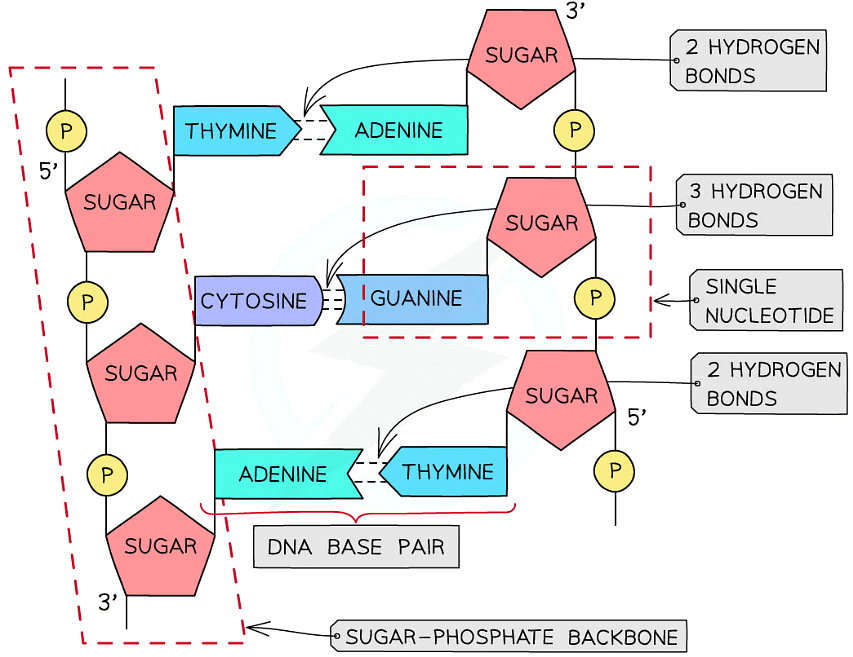
A section of DNA – two antiparallel DNA polynucleotide strands held together by hydrogen bonds
Double helix
- DNA is not two-dimensional as seen in the diagram above
- DNA is described as a double helix
- This refers to the three-dimensional shape that DNA molecules form
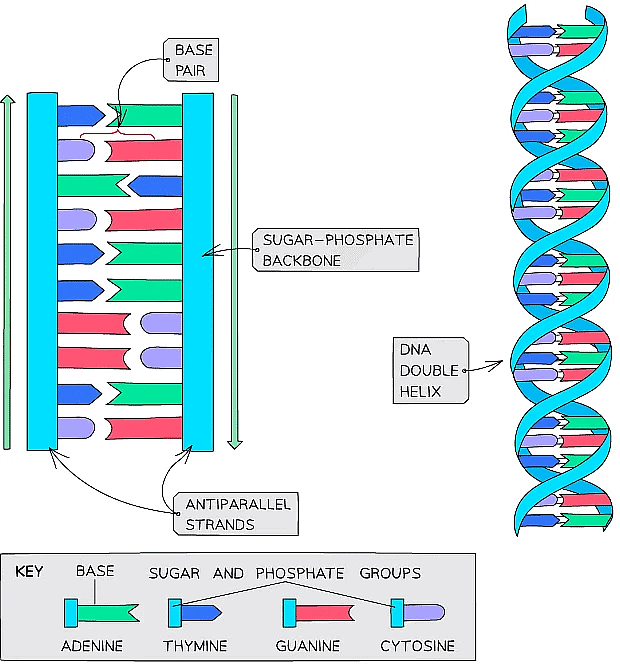
DNA molecules form a three-dimensional structure known as a DNA double helix
The Structure of RNA
- Like DNA, the nucleic acid RNA (ribonucleic acid) is a polynucleotide – it is made up of many nucleotides linked together in a chain
- Like DNA, RNA nucleotides contain the nitrogenous bases adenine (A), guanine (G) and cytosine (C)
- Unlike DNA, RNA nucleotides never contain the nitrogenous base thymine (T) – in place of this they contain the nitrogenous base uracil (U)
- Unlike DNA, RNA nucleotides contain the pentose sugar ribose (instead of deoxyribose)
 An RNA nucleotide compared with a DNA nucleotide
An RNA nucleotide compared with a DNA nucleotide
- Unlike DNA, RNA molecules are only made up of one polynucleotide strand (they are single-stranded)
- RNA polynucleotide chains are relatively short compared to DNA
- Each RNA polynucleotide strand is made up of alternating ribose sugars and phosphate groups linked together, with the nitrogenous bases of each nucleotide projecting out sideways from the single-stranded RNA molecule
- The sugar-phosphate bonds (between different nucleotides in the same strand) are covalent bonds known as phosphodiester bonds
- These bonds form what is known as the sugar-phosphate backbone of the RNA polynucleotide strand
- The phosphodiester bonds link the 5-carbon of one ribose sugar molecule to the phosphate group from the same nucleotide, which is itself linked by another phosphodiester bond to the 3-carbon of the ribose sugar molecule of the next nucleotide in the strand
- An example of an RNA molecule is messenger RNA (mRNA), which is the transcript copy of a gene that encodes a specific polypeptide. Two other examples are transfer RNA (tRNA) and ribosomal RNA (rRNA)
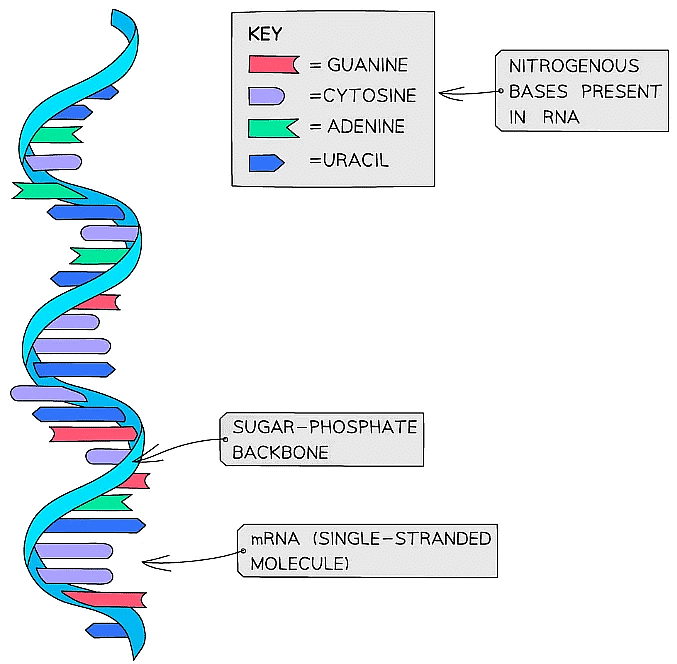 Messenger RNA (mRNA) provides a good example of the structure of RNA
Messenger RNA (mRNA) provides a good example of the structure of RNA
Ribosomes
- Ribosomes are small organelles that are either free in the cytoplasm (of all cells) or are attached to the rough endoplasmic reticulum (only in eukaryotic cells)
- Ribosomes are the site of protein synthesis (where proteins are made)
- They ‘read’ RNA to make polypeptides (proteins) in a process known as translation
- Ribosomes are themselves formed from RNA and proteins
- The RNA that forms part of the structure of ribosomes is a specific type of RNA known as ribosomal RNA (rRNA)
- The rRNA in ribosomes has enzymatic properties that catalyse the formation of peptide bonds between amino acids
- Each ribosome is a mixture of ribosomal RNA and proteins
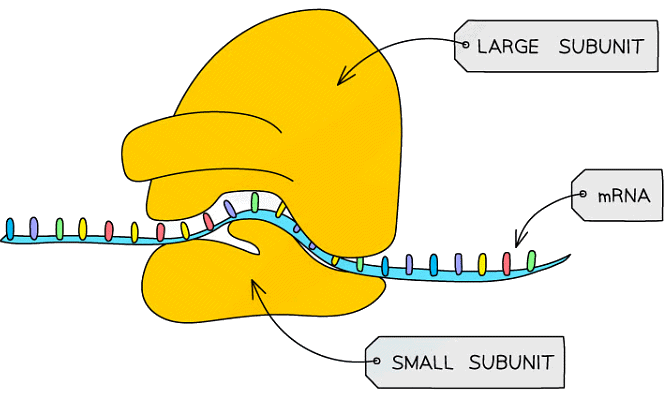
- Ribosomes in eukaryotic cells are larger than those in prokaryotic cells. In both cell types, ribosomes are composed of a small subunit and a large subunit
- 80S ribosomes (composed of 60S and 40S subunits) are found in eukaryotic cells
- 70S ribosomes (composed of 50S and 30S subunits) are found in prokaryotic cells, as well as in the mitochondria and chloroplasts of eukaryotic cells
- The large subunit is the site of translation
- The rRNA and proteins of the large subunit hold tRNA molecules (with their attached amino acids) in place
- rRNA can then catalyse the condensation reactions between amino acids
- mRNA sits between the two subunits and the ribosome moves along it as it translates it into a polypeptide
- Unlike some organelles, ribosomes are not surrounded by a membrane

A diagram of a ribosome, showing the small and large subunits
The Origins of Research on the Genetic Code
Appreciating How the Function of DNA Was Determined
- DNA was actually first observed in the 1800s by a Swiss scientist called Friedrich Miescher
- Miescher is credited with being the first person to discover DNA (although he named it ‘nuclein’) and define it as a distinct molecule in 1869
- However, many scientific researchers at that time doubted that this newly discovered DNA molecule could carry the genetic code
- They doubted this because of the relatively simple chemical composition of DNA (because DNA was only made up of simple repeating nucleotides, which themselves were only composed of three parts: a phosphate group, deoxyribose, a nitrogen-containing organic base
- For example, some scientists hypothesised that genetic information must be carried by proteins, which show much higher levels of chemical complexity
- For example, proteins are which are made up of 20 different amino acids whereas DNA is made up of only 4 different nucleotides
- As a result, it wasn’t until the 1940s that the role of DNA in genetic inheritance began to be more fully researched and understood
- By 1953, experiments had confirmed that DNA carried the genetic code
- It was understood that, despite there being only 4 nucleotides, the use of the triplet code enabled much variation (the code is universal and degenerate)
- The location of DNA, protected in the nucleus, enabled the security of the genetic material rather than proteins that are found in the cytoplasm and susceptible to hydrolysis
- DNA is easily copied and therefore conserved throughout generations of cells and inherited between generations within families
- 1953 was also the year in which Watson and Crick confirmed the double-helix structure of DNA using Rosalind Franklin’s X-ray data
The Purpose of Semi-Conservative Replication
- Before a (parent) cell divides, it needs to copy the DNA contained within it
- This is so that the two new (daughter) cells produced will both receive the full copies of the parental DNA
- The DNA is copied via a process known as semi-conservative replication (semi = half)
- The process is called so because in each new DNA molecule produced, one of the polynucleotide DNA strands (half of the new DNA molecule) is from the original DNA molecule being copied
- The other polynucleotide DNA strand (the other half of the new DNA molecule) has to be newly created by the cell
- Therefore, the new DNA molecule has conserved half of the original DNA and then used this to create a new strand
The importance of retaining one original DNA strand
- It ensures there is genetic continuity between generations of cells
- In other words, it ensures that the new cells produced during cell division inherit all their genes from their parent cells
- This is important because cells in our body are replaced regularly and therefore we need the new cells to be able to do the same role as the old ones
- Replication of DNA and cell division also occurs during growth
The Process of Semi-Conservative Replication
- DNA replication occurs in preparation for mitosis, when a parent cell divides to produce two genetically identical daughter cells – as each daughter cell contains the same number of chromosomes as the parent cell, the number of DNA molecules in the parent cell must be doubled before mitosis takes place
- DNA replication occurs during the S phase of the cell cycle (which occurs during interphase, when a cell is not dividing)
- The enzyme helicase unwinds the DNA double helix by breaking the hydrogen bonds between the base pairs on the two antiparallel polynucleotide DNA strands to form two single polynucleotide DNA strands
- Each of these single polynucleotide DNA strands acts as a template for the formation of a new strand made from free nucleotides that are attracted to the exposed DNA bases by base pairing
- The new nucleotides are then joined together by DNA polymerase which catalyses condensation reactions to form a new strand
- The original strand and the new strand joined together through hydrogen bonding between base pairs to form the new DNA molecule
- This method of replicating DNA is known as semi-conservative replication because half of the original DNA molecule is kept (conserved) in each of the two new DNA molecules
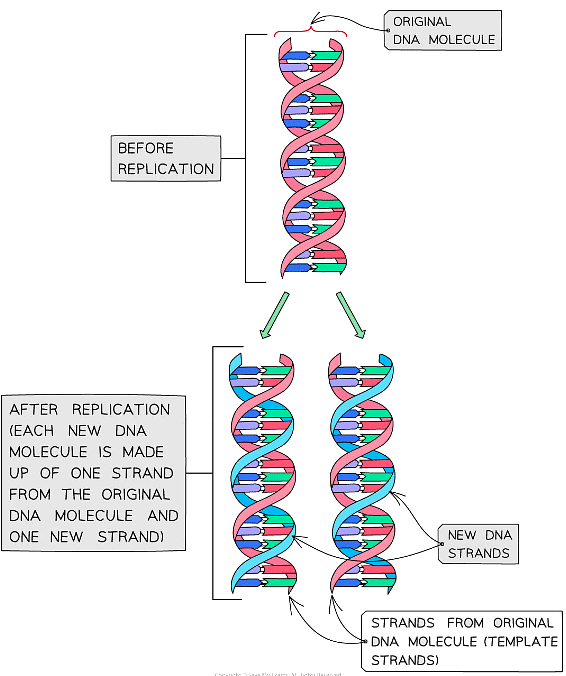
Semi-conservative replication of DNA
DNA Polymerase
- In the nucleus, there are free nucleotides which contain three phosphate groups
- These nucleotides are known as nucleoside triphosphates or ‘activated nucleotides’
- The extra phosphates activate the nucleotides, enabling them to take part in DNA replication
- The bases of the free nucleoside triphosphates align with their complementary bases on each of the template DNA strands
- The enzyme DNA polymerase synthesises new DNA strands from the two template strands
- It does this by catalysing condensation reactions between the deoxyribose sugar and phosphate groups of adjacent nucleotides within the new strands, creating the sugar-phosphate backbone of the new DNA strands
- DNA polymerase cleaves (breaks off) the two extra phosphates and uses the energy released to create the phosphodiester bonds (between adjacent nucleotides)
- Hydrogen bonds then form between the complementary base pairs of the template and new DNA strands
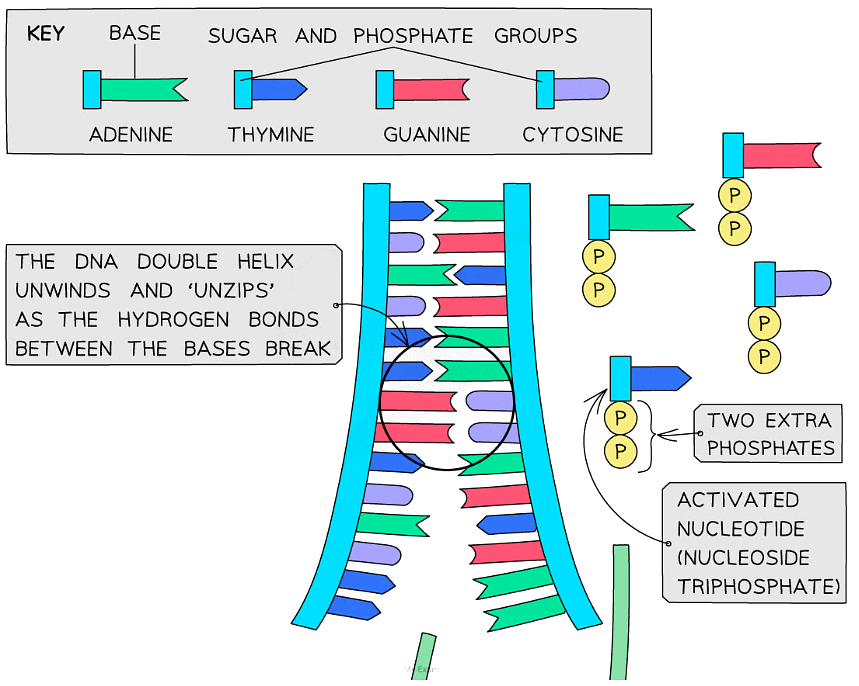
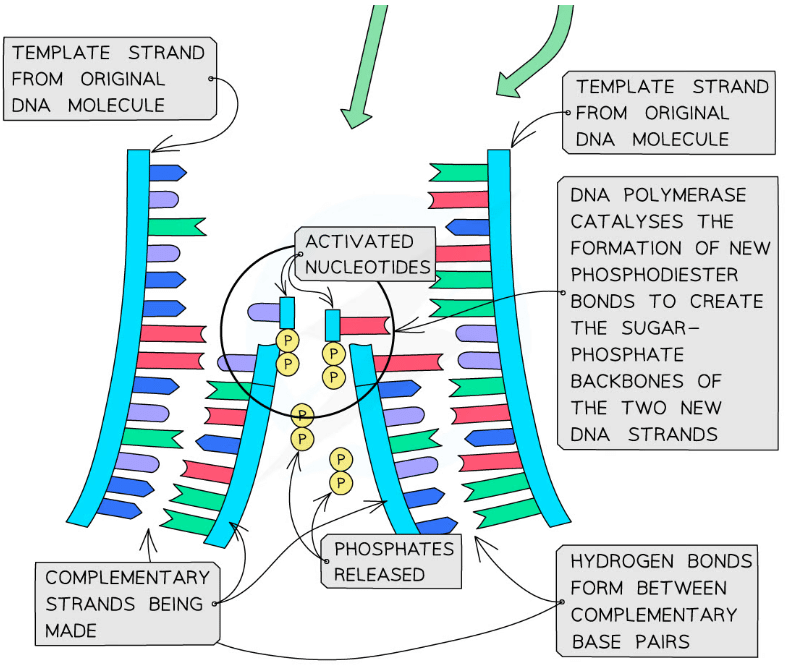
Nucleotides are bonded together by DNA polymerase to create the new complementary DNA strands
Leading & lagging strands
- DNA polymerase can only build the new strand in one direction (5’ to 3’ direction)
- As DNA is ‘unzipped’ from the 3’ towards the 5’ end, DNA polymerase will attach to the 3’ end of the original strand and move towards the replication fork (the point at which the DNA molecule is splitting into two template strands)
- This means the DNA polymerase enzyme can synthesise the leading strand continuously
- This template strand that the DNA polymerase attaches to is known as the leading strand
- The other template strand created during DNA replication is known as the lagging strand
- On this strand, DNA polymerase moves away from the replication fork (from the 5’ end to the 3’ end)
- This means the DNA polymerase enzyme can only synthesise the lagging DNA strand in short segments (called Okazaki fragments)
- A second enzyme known as DNA ligase is needed to join these lagging strand segments together to form a continuous complementary DNA strand
- DNA ligase does this by catalysing the formation of phosphodiester bonds between the segments to create a continuous sugar-phosphate backbone
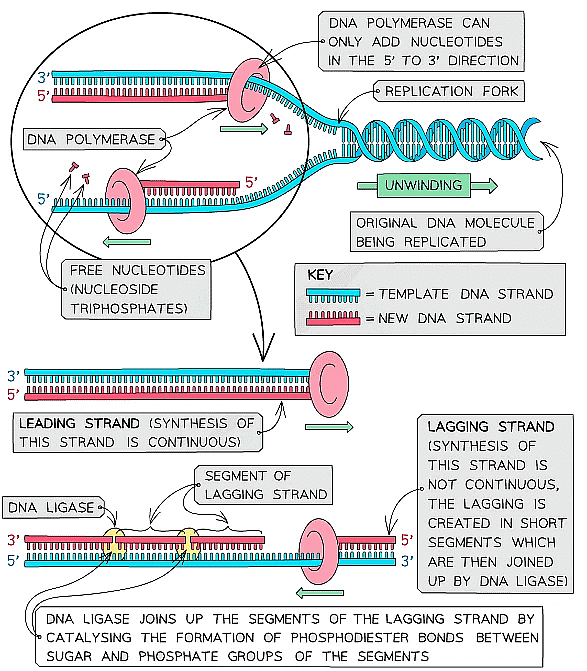
The synthesis of the complementary strands occurs slightly differently on the leading and lagging template strands of the original DNA molecule that is being replicated
Calculating the Frequency of Nucleotide Bases
- All DNA nucleotides contain the same phosphate group and deoxyribose sugar. The nitrogenous base attached to the sugar can vary between nucleotides
- There are four different bases:
- Adenine (A)
- Cytosine (C)
- Thymine (T)
- Guanine (G)
- The bases on each strand pair up with each other, holding the two strands of DNA in a double helix
- The bases always pair up in the same way:
- Adenine always pairs with Thymine (A-T)
- Cytosine always pairs with Guanine (C-G)
- The bases in each of these pairs are said to be complementary to one another
- This means that the frequency or number of adenine is equal to the frequency or number of thymine and the frequency or number of cytosine is equal to the frequency or number of guanine
- In an exam, you could be given the frequency of just one of the bases in a DNA molecule and asked to determine the frequency of the other three bases
Example: In a section of DNA, 28% of the bases were cytosine. Calculate the frequencies of the other three bases.
Step 1: Find the number of guanine bases
- C = G, therefore 28% of the bases must be guanine
- In total, therefore, cytosine and guanine make up 56% of the total bases
Step 2: Find the number of adenine and thymine bases
- This leaves 44% of the bases as adenine and thymine
- A = T, therefore 22% of the bases must be adenine and 22% of the bases must be thymine
The Watson Crick Model
Evaluating the Watson-Crick Model
- Watson and Crick were two scientists who worked together to confirm the double-helix structure of DNA in 1953
- Watson and Crick also came up with a model by which DNA might be replicated:
- This theory was called semi-conservative DNA replication
- The theory is based upon the specific hydrogen bonding between pairs of nitrogenous bases (A+T and C+G) being used during replication to conserve the genetic sequence
- However, this was just a theory, another theory suggested DNA replicated ‘conservatively’
- The theory of conservative DNA replication suggested that the strands of the original DNA molecule would stay together, and the new, replicated DNA molecule would be made out of two brand new strands
- Watson and Crick’s theory of semi-conservative DNA replication was later proved to be correct by the work of two other scientists, Meselson and Stahl
- They used bacteria and two nitrogen isotopes, a heavy form (15N) and the normal, lighter form (14N), to prove this
Meselson and Stahl’s Experiment
- Bacteria are grown in a broth containing the heavy (15N) nitrogen isotope
- DNA contains nitrogen in its bases
- As the bacteria replicated, they used nitrogen from the broth to make new DNA nucleotides
- After some time, the culture of bacteria had DNA containing only heavy (15N) nitrogen
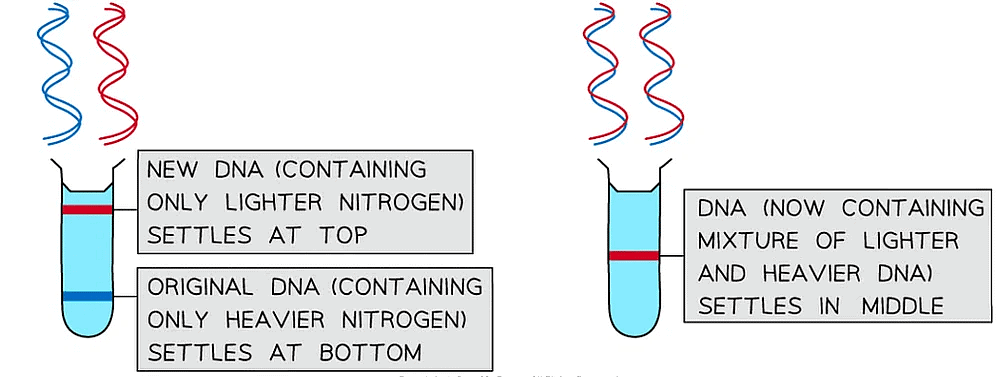
- A sample of DNA from the 15N culture of bacteria was extracted and spun in a centrifuge
- This showed that the DNA containing the heavy nitrogen settled near the bottom of the centrifuge tube
- The bacteria containing only 15N DNA was then taken out of the 15N broth and added to a broth containing only the lighter 14N nitrogen. The bacteria were left for enough time for one round of DNA replication to occur before their DNA was extracted and spun in a centrifuge
- If conservative DNA replication had occurred, the original template DNA molecules would only contain the heavier nitrogen and would settle at the bottom of the tube, whilst the new DNA molecules would only contain the lighter nitrogen and would settle at the top of the tube
- If semi-conservative replication had occurred, all the DNA molecules would now contain both the heavy 15N and light 14N nitrogen and would therefore settle in the middle of the tube (one strand of each DNA molecule would be from the original DNA containing the heavier nitrogen and the other (new) strand would be made using only the lighter nitrogen)
- Meselson and Stahl confirmed that the bacterial DNA had undergone semi-conservative replication.
- The DNA from this second round of centrifugation settled in the middle of the tube, showing that each DNA molecule contained a mixture of the heavier and lighter nitrogen isotopes
- If more rounds of replication were allowed to take place, the ratio of 15N:14N would go from 1:1 after the first round of replication, to 3:1 after the second and 7:1 after the third
- This experiment proved Watson and Crick's theory correct



Meselson and Stahl’s experiment that showed bacterial DNA replicated via semi-conservative DNA replication
|
122 videos|161 docs|138 tests
|

|
Explore Courses for Grade 12 exam
|

|