Processes of Recombinant DNA Technology | Biology Class 12 - NEET PDF Download
Introduction
Recombinant DNA (rDNA) technology refers to the process of joining DNA molecules from two different sources and inserting them into a host organism, to generate products for human use. Can you put the DNA molecules in the host organism first and then cut and join them? No! This process involves multiple steps that have to proceed in a specific sequence to generate the desired product.
Steps of Recombinant DNA Technology
Let’s understand each step in detail.
1. Isolation of Genetic Material
We already know that the genetic material of all living organisms is ‘nucleic acid’. In most organisms, it is DNA, whereas in some it is RNA. The first step in rDNA technology is to isolate the desired DNA in its pure form i.e. free from other macromolecules.
However, in a normal cell, the DNA not only exists within the cell membrane, but is also present along with other macromolecules such as RNA, polysaccharides, proteins, and lipids. So, how do we break open the cell and obtain DNA that is free from other macromolecules? We can use the following enzymes for specific purposes:
- Lysozyme – to break bacterial cell wall.
- Cellulase – to break plant cell wall.
- Chitinase – to break fungal cell wall.
- Ribonuclease – removes RNA.
- Protease – removes proteins (such as histones that are associated with DNA).
Other macromolecules are removable with other enzymes or treatments. Ultimately, the addition of ethanol causes the DNA to precipitate out as fine threads. This is then spooled out to give purified DNA.
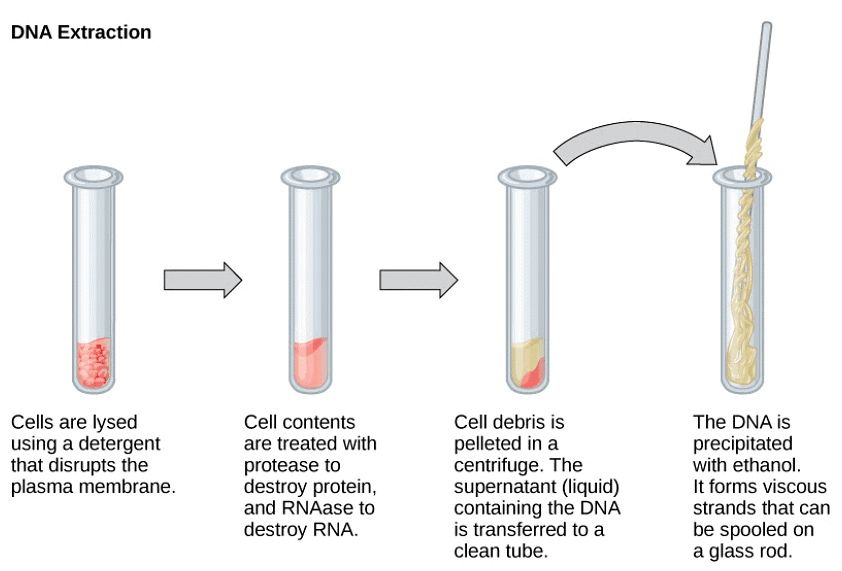 Extraction of DNA
Extraction of DNA
2. Restriction Enzyme Digestion
Restriction enzymes act as molecular scissors that cut DNA at specific locations. These reactions are called ‘restriction enzyme digestions’. They involve the incubation of the purified DNA with the selected restriction enzyme, at conditions optimal for that specific enzyme.
The technique – ‘Agarose Gel Electrophoresis’ reveals the progress of the restriction enzyme digestion. This technique involves running out the DNA on an agarose gel. On the application of current, the negatively charged DNA travels to the positive electrode and is separated out based on size. This allows us to separate and cut out the digested DNA fragments. The vector DNA is also processed using the same procedure.
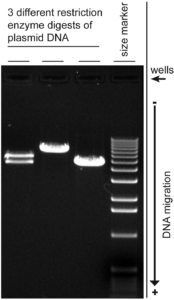 Digested DNA Electrophoresis
Digested DNA Electrophoresis
3. Amplification using PCR
Polymerase Chain Reaction or PCR is a method of making multiple copies of a DNA sequence using the enzyme – DNA polymerase. It helps to amplify a single copy or a few copies of DNA into thousands to millions of copies. PCR reactions are run on ‘thermal cyclers’ using the following components:
- Template – DNA to be amplified
- Primers – small, chemically synthesized oligonucleotides that are complementary to a region of the DNA.
- Enzyme – DNA polymerase
- Nucleotides – needed to extend the primers by the enzyme.
 Thermal Cycler
Thermal Cycler
The cut fragments of DNA can be amplified using PCR and then ligated with the cut vector.
Polymerase Chain Reaction Technology (PCR - Technology)
- This technique was invented by Kary Mullis (1983).
- In 1993 Karry Mullis got a Nobel prize for PCR(for chemistry)
- PCR is a method for amplifying a specific region of DNA molecule without the requirement for time-consuming cloning procedures. – PCR reaction takes place in the Eppendorf tube.
- Using PCR-technique very low content of DNA available from samples of blood or semen or any other tissue or hair cell can be amplified many times and analysed. In this technique, Taq-Polymerase is used.
- Taq polymerase enzyme is used in PCR which is a special type of DNA polymerase enzyme which is resistant to high temperature. Taq Polymerase is isolated from the Thermus aquaticus bacterium.
- Some other examples of polymerase which are used in PCR are
- Pfu Polymerase - Isolated from Pyrococus furiosus bacterium.
- Vent Polymerase- Isolated from Thermococcus litoralis bacterium.
Three Main Steps in PCR
- Denaturation
- Annealing
- Extension
Polymerase Chain Reaction Cycle
- Denaturation: In this step, a double-stranded DNA molecule is placed at 94°C. So double-stranded DNA becomes single-stranded & each single-stranded DNA functions as a template.
- Annealing: In this step, two primer DNA are attached at the 3' end of single-stranded DNA.
- Extension: In this process, the Taq polymerase enzyme synthesizes DNA strain over the template. PCR is an automatic process because taq. polymerase enzyme is heat resistant.
4. Ligation of DNA Molecules
The purified DNA and the vector of interest are cut with the same restriction enzyme. This gives us the cut fragment of DNA and the cut vector, that is now open. The process of joining these two pieces together using the enzyme ‘DNA ligase’ is ‘ligation’. The resulting DNA is ‘recombinant DNA‘.
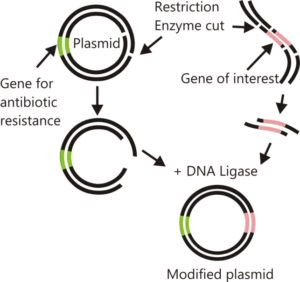 Restriction Enzyme Digestion followed by Ligation
Restriction Enzyme Digestion followed by Ligation
5. Insertion of Recombinant DNA into Host
In this step, the recombinant DNA is introduced into a recipient host cell. This process is ‘Transformation’. Bacterial cells do not accept foreign DNA easily. Therefore, they are treated to make them ‘competent’ to accept new DNA. (The topic – Tools of Biotechnology explains a few ways to make cells competent).
During transformation, if a recombinant DNA bearing a gene for ampicillin resistance is transferred into recipient E. coli cells, then the E. coli cells also become ampicillin-resistant. This aspect is useful in differentiating transformed cells from non-transformed cells.
For example, if we spread the transformed cells on agar plates containing ampicillin, only the transformed, ampicillin-resistant cells will grow while the untransformed cells will die. Therefore, in this case, the ampicillin resistance gene acts as the ‘selectable marker’.
6. Obtaining Foreign Gene Product
The recombinant DNA multiplies in the host and is expressed as a protein, under optimal conditions. This is now a recombinant protein. Small volumes of cell cultures will not yield a large amount of recombinant protein. Therefore, large-scale production is necessary to generate products that benefit humans. For this purpose, vessels called bioreactors are used.
Bioreactors are large containers with a continuous culture system, where the fresh medium is added from one side and used medium is taken out from another side. Bioreactors can process about 100-1000 litres of cell cultures. A bioreactor provides optimum conditions (temperature, oxygen, pH, vitamins etc.) to biologically convert raw materials into specific proteins, enzymes etc.
‘Stirred-tank bioreactor’ is the most common type of bioreactor. It is usually cylindrical and has the following parts:
- Agitator system – to stir the contents evenly
- Oxygen delivery system – to introduce air into the system
- Foam control system
- Temperature control system
- pH control system
- Sampling ports – to take out small amounts of culture
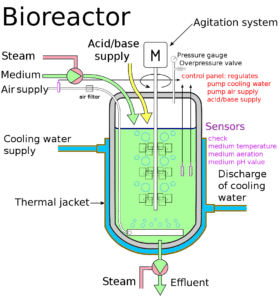 Bioreactor
Bioreactor
7. Downstream Processing
Before the protein is marketed as a final product, it is subjected to downstream processing which includes:
- Separation and purification.
- Formulation with suitable preservatives.
- Clinical trials to test the efficacy and safety of the product.
- Quality control tests.
|
78 videos|386 docs|202 tests
|
FAQs on Processes of Recombinant DNA Technology - Biology Class 12 - NEET
| 1. What is recombinant DNA technology? |  |
| 2. What are the steps involved in recombinant DNA technology? |  |
| 3. What are the processes involved in recombinant DNA technology? |  |
| 4. What are the potential applications of recombinant DNA technology? |  |
| 5. What are the ethical concerns surrounding recombinant DNA technology? |  |
















