Tools of Recombinant DNA Technology | Biology Class 12 - NEET PDF Download
| Table of contents |

|
| Tools of Recombinant DNA Technology |

|
| 1. Restriction Enzymes |

|
| 2. Cloning Vectors |

|
| 3. Competent Host |

|
Tools of Recombinant DNA Technology
- Recombinant DNA technology, also known as genetic engineering, relies on key tools to manipulate and transfer genetic material. These tools include:
- Restriction Enzymes
- Polymerase Enzymes
- Ligases
- Vectors
- Host Organism
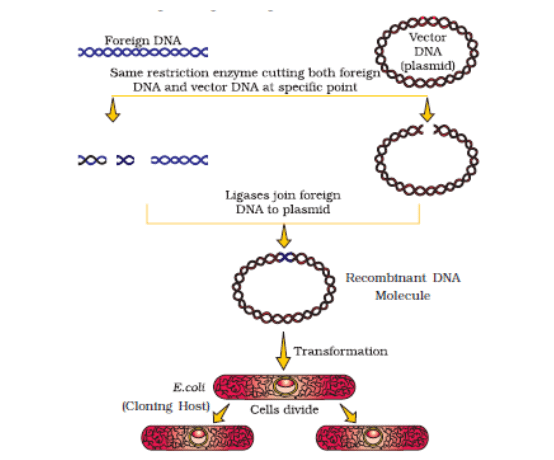 Diagrammatic representation of recombinant DNA technology
Diagrammatic representation of recombinant DNA technology
Let's explore each of these tools in detail, starting with restriction enzymes.
1. Restriction Enzymes
Restriction enzymes, also known as restriction endonucleases, are specialized proteins that cut DNA at specific sequences. They play a crucial role in genetic engineering by allowing scientists to precisely cut and manipulate DNA molecules.
- The discovery of restriction enzymes dates back to 1963 when researchers isolated two enzymes from Escherichia coli (E. coli) that restricted the growth of bacteriophages (viruses that infect bacteria). One enzyme added methyl groups to DNA, while the other cut DNA. The cutting enzyme was later named restriction endonuclease.
- One of the first restriction endonucleases, Hind II, was characterized in 1968. Hind II was found to cut DNA at a specific location by recognizing a unique sequence of six base pairs. This specific sequence is known as the recognition sequence for Hind II.
- Today, over 900 different restriction enzymes have been identified, each isolated from various strains of bacteria. These enzymes recognize different DNA sequences and cut the DNA at specific points.
Restriction enzymes are part of a larger group of enzymes called nucleases, which are classified into two types:
(i) Exonucleases: These enzymes remove nucleotides from the ends of a DNA molecule.
(ii) Endonucleases: These enzymes make cuts within the DNA molecule at specific positions.
- Restriction endonucleases work by scanning the DNA sequence for their specific recognition site. When they find their target sequence, they bind to the DNA and cut both strands of the double helix at specific points in the sugar-phosphate backbone.
- Many restriction enzymes recognize palindromic sequences in DNA. A palindromic sequence is one that reads the same forwards and backwards on both strands of DNA. For example, the sequence: 5'—GAATTC—3'3'—CTTAAG—5' is palindromic because it reads the same on both strands from the 5' to the 3' direction.
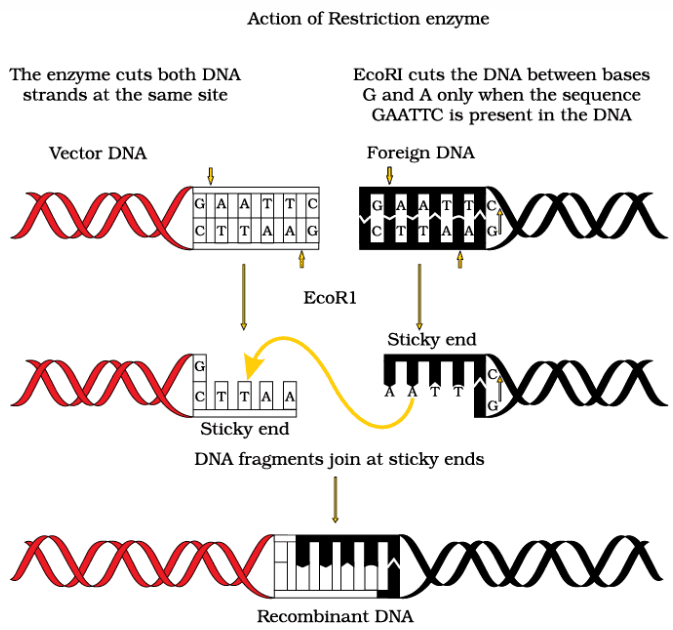 Steps in formation of recombinant DNA by action of restriction endonuclease enzyme - EcoRI
Steps in formation of recombinant DNA by action of restriction endonuclease enzyme - EcoRI
- When restriction enzymes cut DNA, they often do so a little away from the center of the palindromic site, leaving behind single-stranded overhangs known as sticky ends. These sticky ends can form hydrogen bonds with complementary sticky ends, facilitating the joining of DNA fragments.
- In genetic engineering, restriction endonucleases are used to create recombinant DNA molecules by cutting DNA from different sources and joining them together. When DNA fragments are cut with the same restriction enzyme, they have compatible sticky ends and can be ligated (joined) together using DNA ligase.
- To separate and isolate DNA fragments generated by restriction endonucleases, a technique called gel electrophoresis is used. DNA fragments are negatively charged and move towards the positive electrode (anode) under an electric field through a gel matrix, usually made of agarose. Smaller fragments move farther through the gel than larger ones.
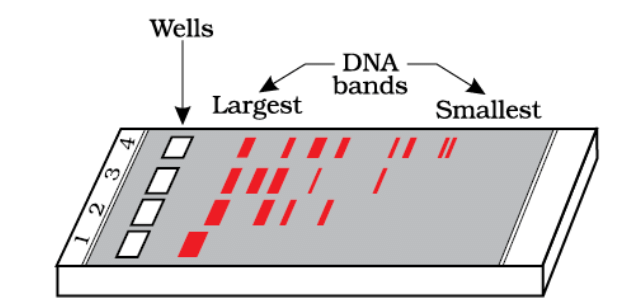 A typical agarose gel electrophoresis showing migration of undigested (lane 1) and digested set of DNA fragments (lane 2 to 4)
A typical agarose gel electrophoresis showing migration of undigested (lane 1) and digested set of DNA fragments (lane 2 to 4)
- The separated DNA fragments are stained with a compound called ethidium bromide and visualized under ultraviolet (UV) light. The bright orange bands seen in the gel represent the separated DNA fragments. These fragments can be cut out from the gel and purified for use in constructing recombinant DNA by joining them with cloning vectors.
2. Cloning Vectors
Vectors are essential tools in molecular biology, designed to facilitate the cloning and propagation of foreign DNA fragments within host cells. Among the most commonly used vectors are plasmids and bacteriophages, which possess the remarkable ability to replicate independently of chromosomal DNA within bacterial cells.
1. Plasmids: Plasmids are circular, double-stranded DNA molecules that can exist independently of the bacterial chromosomal DNA. They can vary in copy number, with some plasmids replicating in high numbers (15-100 copies per cell), while others may have only one or two copies. The copy number is determined by the origin of replication (ori) present in the plasmid. By linking foreign DNA to a plasmid, researchers can ensure that the foreign DNA is replicated and passed on to daughter cells in high quantities.
2. Bacteriophages: Bacteriophages, or phages, are viruses that infect bacteria. They have a high copy number of their genome within bacterial cells due to their ability to produce many progeny. By attaching foreign DNA to bacteriophage DNA, researchers can achieve high levels of replication and propagation of the foreign DNA within bacterial hosts.
Essential Features of Cloning Vectors
Cloning vectors are engineered with specific features to facilitate the easy insertion of foreign DNA and the selection of recombinant clones from non-recombinant ones. The following are the key features required in a cloning vector:
(i) Origin of Replication (ori): The ori is a crucial sequence that determines where replication starts. Any DNA linked to this sequence can replicate within the host cells. The ori also controls the copy number of the linked DNA. To obtain many copies of the target DNA, it is essential to use a vector with a high copy number ori.
(ii) Selectable Marker: A selectable marker is necessary to identify and eliminate non-transformants while allowing the growth of transformants. Common selectable markers include antibiotic resistance genes such as ampicillin, chloramphenicol, tetracycline, or kanamycin. Since normal E. coli cells lack resistance to these antibiotics, only transformed cells carrying the selectable marker can grow in the presence of the antibiotic.
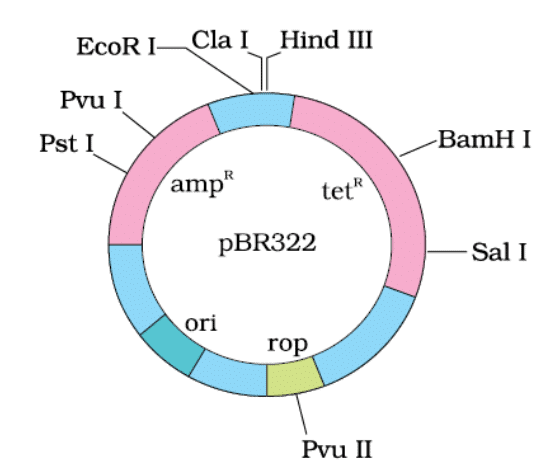 E. coli cloning vector pBR322 showing restriction sites (Hind III, EcoR I, BamH I, Sal I, Pvu II, Pst I, Cla I), ori and antibiotic resistance genes (ampR and tetR ). rop codes for the proteins involved in the replication of the plasmid.
E. coli cloning vector pBR322 showing restriction sites (Hind III, EcoR I, BamH I, Sal I, Pvu II, Pst I, Cla I), ori and antibiotic resistance genes (ampR and tetR ). rop codes for the proteins involved in the replication of the plasmid.
(iii) Cloning Sites
- The vector should have minimal, preferably single, recognition sites for commonly used restriction enzymes. Having multiple recognition sites can complicate gene cloning by generating several fragments. The foreign DNA is ligated at a restriction site within one of the antibiotic resistance genes. For example, in the vector pBR322, foreign DNA can be ligated at the BamH I site of the tetracycline resistance gene.
- When a recombinant plasmid is formed, it loses tetracycline resistance due to the insertion of foreign DNA. However, it can still be selected from non-recombinant plasmids by plating transformants on tetracycline-containing medium. Transformants growing on ampicillin-containing medium are then transferred to a medium containing tetracycline. The recombinants will grow on ampicillin but not on tetracycline, while non-recombinants will grow on both media.
- Selection of recombinants based on antibiotic resistance can be cumbersome as it requires simultaneous plating on two different antibiotic media. To simplify this process, alternative selectable markers have been developed, such as using chromogenic substrates to differentiate recombinants from non-recombinants based on their ability to produce color. Insertional inactivation of the β-galactosidase gene is one such method, where the presence of a chromogenic substrate produces blue colonies if the plasmid lacks an insert, and colorless colonies if the plasmid has an insert.
(iv) Vectors for Cloning Genes in Plants and Animals
Vectors designed for cloning genes in plants and animals have been inspired by the natural gene delivery mechanisms employed by pathogens like bacteria and viruses. These vectors facilitate the introduction of foreign genes into eukaryotic cells, enabling the desired genetic modifications.
1. Vectors for Plant Gene Cloning: One of the most well-known methods for delivering genes into plants involves the use of the tumor-inducing (Ti) plasmid from Agrobacterium tumefaciens, a bacterium that causes tumors in certain dicot plants. The Ti plasmid has been modified into a cloning vector that is no longer pathogenic but retains its ability to deliver genes of interest into a variety of plants. This process takes advantage of the natural mechanisms used by the bacterium to transfer DNA into plant cells, allowing for the stable integration of foreign genes.
2. Vectors for Animal Gene Cloning: Similarly, retroviruses, which are known for their ability to induce cancer by integrating viral DNA into the host cell's genome, have also been repurposed as gene delivery vectors for animal cells. These retroviral vectors have been disarmed to remove their pathogenic properties while retaining the capacity to introduce and integrate desired genes into the genomes of animal cells. This technology has proven valuable for various applications, including gene therapy and genetic engineering in animals.
Once a gene or DNA fragment is ligated into a suitable vector, the vector is introduced into a bacterial, plant, or animal host, where it can multiply and express the inserted genetic material. This process allows researchers to manipulate and study genes in different organisms, paving the way for advances in genetics, agriculture, and medicine.
3. Competent Host
Why DNA Cannot Pass Through Cell Membranes: DNA is a hydrophilic (water-attracting) molecule, which is why it cannot easily pass through the hydrophobic (water-repelling) lipid bilayer of cell membranes. To facilitate the uptake of plasmid DNA by bacterial cells, these cells must be made 'competent.' This is typically achieved by treating the bacterial cells with a specific concentration of a divalent cation, such as calcium. This treatment increases the efficiency with which DNA enters the bacterium through pores in its cell wall.
Methods of Introducing Recombinant DNA into Host Cells:
- Heat Shock Method: Once the bacterial cells are made competent, recombinant DNA can be introduced by incubating the cells with the DNA on ice. This is followed by a brief exposure to a higher temperature (around 42°C) to create a heat shock, after which the cells are put back on ice. This process helps the bacteria take up the recombinant DNA.
- Micro-Injection: In this method, recombinant DNA is directly injected into the nucleus of an animal cell using a fine needle. This technique allows for precise delivery of DNA into the target cells.
- Biolistics or Gene Gun: This method is suitable for plant cells. In biolistics, high-velocity micro-particles made of gold or tungsten, coated with DNA, are bombarded into the plant cells. This physical method facilitates the introduction of DNA into the cells.
- Disarmed Pathogen Vectors: In this approach, modified pathogens are used to transfer recombinant DNA into the host cells. These disarmed pathogens infect the cells and deliver the recombinant DNA as part of their infection process.
|
59 videos|290 docs|168 tests
|
FAQs on Tools of Recombinant DNA Technology - Biology Class 12 - NEET
| 1. What are restriction enzymes and how do they function in recombinant DNA technology? |  |
| 2. What are cloning vectors and what roles do they play in recombinant DNA technology? |  |
| 3. What is a competent host and why is it important in recombinant DNA technology? |  |
| 4. How do researchers create competent host cells for transformation? |  |
| 5. What are some applications of recombinant DNA technology in medicine and agriculture? |  |















