Search for Genetic Material | Biology Class 12 - NEET PDF Download
| Table of contents |

|
| The Search for Genetic Material |

|
| Transforming Principle |

|
| The Genetic Material is DNA |

|
| Properties of Genetic Material (DNA versus RNA) |

|
The Search for Genetic Material
- Despite the simultaneous discovery of nuclein by Meischer and the principles of inheritance by Mendel, it took a considerable time to establish and prove that DNA serves as the genetic material.
- By 1926, the investigation into the mechanisms of genetic inheritance had delved into the molecular realm. Previous findings by notable scientists such as Gregor Mendel, Walter Sutton, and Thomas Hunt Morgan had focused the inquiry on chromosomes within the nucleus of most cells. However, the specific molecule responsible for genetic material remained unidentified.
Transforming Principle
- In 1928, Frederick Griffith conducted experiments with Streptococcus pneumoniae, the bacterium responsible for pneumonia, and observed a remarkable transformation in the bacteria.
- During his experiments, Griffith witnessed a change in the physical form of the bacteria. When S. pneumoniae(pneumococcus) bacteria are cultivated on a culture plate, some strains produce smooth, shiny colonies (S), while others produce rough colonies (R).
- The S strain bacteria have a mucous (polysaccharide) coat, whereas the R strain lacks this coat. Mice infected with the S strain (virulent) succumb to pneumonia, while those infected with the R strain do not develop the disease.
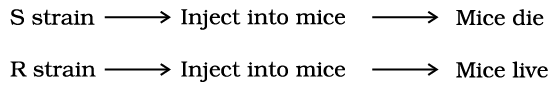
- Griffith discovered that he could kill bacteria by heating them. He noted that mice injected with heat-killed S strain bacteria did not die.
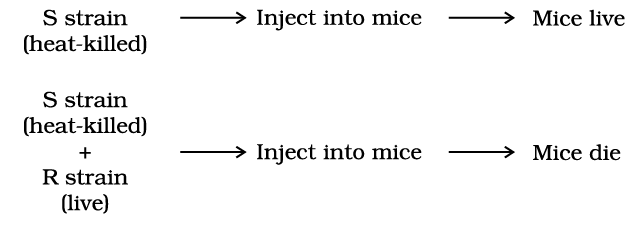
- However, when he injected a mixture of heat-killed S and live R bacteria, the mice died, and he was able to recover living S bacteria from the deceased mice.
- Griffith concluded that the R strain bacteria had been transformed by the heat-killed S strain bacteria. Some “transforming principle,” transferred from the heat-killed S strain, had enabled the R strain to produce a smooth polysaccharide coat and become virulent.
- This suggested the transfer of genetic material. However, Griffith's experiments did not define the biochemical nature of the genetic material.
Biochemical Characterization of Transforming Principle
Before the work of Oswald Avery, Colin MacLeod, and Maclyn McCarty(1933-44), the prevailing thought was that the genetic material was a protein. These researchers aimed to determine the biochemical nature of the “transforming principle” in Griffith's experiment. They purified various biochemicals (proteins, DNA, RNA, etc.) from the heat-killed S cells to identify which ones could transform live R cells into S cells.
Their findings revealed that only DNA from S bacteria could transform R bacteria into S bacteria. Additionally, they discovered that protein-digesting enzymes (proteases) and RNA-digesting enzymes (RNases) did not affect the transformation, indicating that the transforming substance was neither a protein nor RNA. The use of DNase, which inhibits transformation, suggested that DNA was responsible for the transformation.
Avery, MacLeod, and McCarty concluded that DNA is the hereditary material. However, not all biologists were immediately convinced by their findings.
The Genetic Material is DNA
Experiments conducted by Alfred Hershey and Martha Chase in 1952 provided clear evidence that DNA is the genetic material. They focused on bacteriophages, which are viruses that infect bacteria. These viruses attach to bacteria and inject their genetic material into the bacterial cell, which then uses this material to produce more virus particles.
 Hershey- Chase Experiment
Hershey- Chase Experiment
Hershey and Chase's Experiment
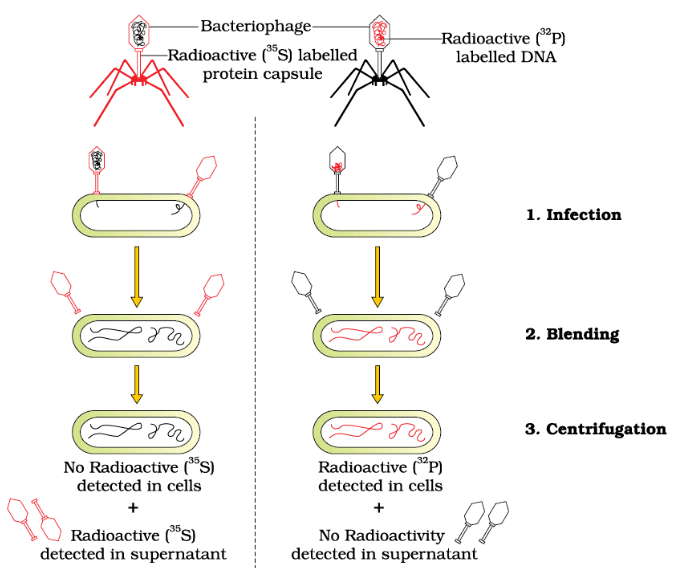
- Hershey and Chase aimed to determine whether it was the protein or the DNA of the viruses that entered the bacteria.
- They grew some viruses in a medium containing radioactive phosphorus and others in a medium with radioactive sulfur.
- Viruses grown with radioactive phosphorus had radioactive DNA but not radioactive protein because DNA contains phosphorus, while protein does not.
- Conversely, viruses grown with radioactive sulfur had radioactive protein but not radioactive DNA, as DNA does not contain sulfur.
Process
- Radioactive phages were allowed to attach to E. coli bacteria.
- During the infection, the viral coats were removed from the bacteria by blending the mixture.
- The virus particles were separated from the bacteria by centrifugation.
Results
- Bacteria infected with viruses containing radioactive DNA were radioactive, indicating that DNA was the material transferred from the virus to the bacteria.
- Bacteria infected with viruses containing radioactive proteins were not radioactive, showing that proteins did not enter the bacteria.
Conclusion
These experiments demonstrated that DNA, not protein, is the genetic material passed from virus to bacteria.
 |
Download the notes
Search for Genetic Material
|
Download as PDF |
Properties of Genetic Material (DNA versus RNA)
From the previous discussion, it is evident that the debate over whether proteins or DNA serve as the genetic material was definitively settled by the Hershey-Chase experiment, establishing DNA as the genetic material. However, it was later discovered that in certain viruses, such as Tobacco Mosaic viruses and QB bacteriophage, RNA functions as the genetic material. The question of why DNA is the predominant genetic material while RNA serves dynamic roles as messenger and adapter can be addressed by examining the chemical differences between the two nucleic acids.
Criteria for Genetic Material
- Replication: The molecule must be capable of replicating itself.
- Chemical and Structural Stability: It should be stable both chemically and structurally.
- Mutation: There should be potential for gradual changes (mutations) necessary for evolution.
- Mendelian Characters: It should be able to express itself in the form of Mendelian traits.
Comparison of DNA and RNA as Genetic Material
- Both DNA and RNA can direct their own duplication due to the rules of base pairing and complementarity.
- Proteins do not fulfill the first criterion of being able to replicate themselves.
- Genetic material must be stable throughout different life cycle stages, ages, and physiological changes.
- The stability of genetic material was evident in Griffith’s ‘transforming principle,’ where heat killed bacteria but did not destroy some properties of genetic material.
- The two strands of DNA, being complementary, can rejoin if separated by heat under appropriate conditions.
- The presence of the 2'-OH group in every RNA nucleotide makes RNA reactive, labile, and easily degradable.
- RNA is also known to be catalytic and therefore more reactive.
- DNA is chemically less reactive and structurally more stable than RNA.
- The presence of thymine instead of uracil in DNA adds to its stability.
Mutability and Stability
- Both DNA and RNA can mutate, but RNA, being less stable, mutates at a faster rate.
- Viruses with RNA genomes, due to their shorter lifespan, mutate and evolve more rapidly.
- RNA can directly code for protein synthesis, allowing for easier expression of traits.
- DNA relies on RNA for protein synthesis, and the protein-synthesizing machinery has evolved around RNA.
Conclusion: Both RNA and DNA can function as genetic material, but DNA is preferred for the storage of genetic information due to its greater stability. RNA is better suited for the transmission of genetic information.
|
49 videos|271 docs|174 tests
|
FAQs on Search for Genetic Material - Biology Class 12 - NEET
| 1. What is the structure of DNA and how does it function as genetic material? |  |
| 2. What are the key steps involved in the mechanism of DNA replication? |  |
| 3. How does transcription occur and what is its significance in gene expression? |  |
| 4. What is the lac operon and how does it regulate gene expression in bacteria? |  |
| 5. What was the Human Genome Project and what were its main achievements? |  |