NCERT Exemplar: Molecular Basis of Inheritance - 2 | Biology Class 12 - NEET PDF Download
SHORT ANSWER TYPE QUESTIONS
Q.1. Define transformation in Griffith's experiment. Discuss how it helps in the identification of DNA as the genetic material.
Ans. Transforming Principle
• In 1928, Frederick Griffith, in a series of experiments with Streptococcus pneumoniae (bacterium responsible for pneumonia), witnessed a miraculous transformation, in the bacteria. During the course of his . experiment, a living organism (bacteria) had changed in physical form.
• When Streptococcus pneumoniae (pneumococcus) bacteria are grown on a culture plate, some produce smooth shiny colonies (S) while others produce rough colonies (R).
• This is because the S strain bacteria have a mucous (polysaccharide) coat, while R strain does not. Mice infected with the S strain (virulent) die from pneumonia infection but mice infected with the R strain do not develop pneumonia. .
S strain -»Inject into mice -» Mice die R strain —»Inject into mice —> Mice live
• Griffith was able to kill bacteria by heating them. He observed that heat- killed S strain bacteria injected into mice did not kill them.
S strain (heat killed) —> Inject into mice —> Mice live
• When he injected a mixture of heat-killed S and live R bacteria, the mice died. Moreover, he recovered living S bacteria from the dead mice.
S strain (heat killed ) + —> Inject into mice —> Mice die R strain (live)
• He concluded that the R strain bacteria had somehow been transformed by the heat-killed S strain bacteria. Some ‘transforming principle’, transferred from the heat-killed S strain, had enabled the R strain to synthesise a smooth polysaccharide coat and become virulent. This must be due to the transfer of the genetic material. However, the biochemical nature of genetic material was not defined from his experiments.
Q.2. Who revealed biochemical nature of the transforming principle? How was it done?
Ans. Prior to the work of Oswald Avery, Colin MacLeod and Maclyn McCarty (1933-44), the genetic material was thought to be a protein. They worked th determine the biochemical nature of ‘transforming principle’ in Griffith’s experiment. They purified biochemicals (proteins, DNA, RNA, etc.) from the heat-killed S cells to see which ones could transform live R cells into S cells. They discovered that DNA alone from S bacteria caused R bacteria to become transformed.
They also discovered that protein-digesting enzymes (proteases) and RNA-digesting enzymes (RNases) did not affect transformation, so the transforming substance was not a protein or RNA. Digestion with DNase did inhibit transformation, suggesting that the DNA caused the transformation. They concluded that DNA is the hereditary material, but not all biologists were convinced.
Q.3. Discuss the significance of heavy isotope of nitrogen in the Meselson and Stahl’s experiment.
Ans. Matthew Meselson and Franklin Stahl performed the following experiment in 1958. They grew E. coli in a medium containing 15NH4C1 (15N is the heavy isotope of nitrogen) as the only nitrogen source for many generations. The result was that l5N was incorporated into newly synthesised DNA (as well as other nitrogen containing compounds).
This heavy DNA molecule could be .distinguished from the normal DNA by centrifugation in a cesium chloride (CsCl) density gradient 15N is not a radioactive isotope, and it can be separated from 14N only based on densities).
Q.4. Define a cistron. Giving examples differentiate between monocistronic and polyeistronic transcription unit.
Ans. Portion of DNA having information for an entire polypeptide or trait is called cistron. However by defining a cistron as a segment of DNA coding for a polypeptide, the structural gene in a transcription unit jcould be said as monocistronic (mostly in eukaryotes) or polycistronic (mostly in bacteria or prokaryotes). In eukaryotes, the monocistronic structural genes have interrupted coding sequences—the genes in eukaryotes are split.
The coding sequences or expressed sequences are defined as exons. Exons are said to be those sequence that appear in mature or processed RNA. The exons are interrupted by introns. Introns or intervening sequences do not appear in mature or processed RNA.
Q.5. Give any six features of the human genome.
Ans. Some of the salient Observations drawn from human genome project are as follows:
1. The human genome contains 3164.7 million nucleotide bases.
2. The average gene consists of 3000 bases, but sizes vary greatly, with the largest known human gene being dystrophin at 2.4 million bases.
3. The total number of genes is estimated at 30,000-much lower than previous estimates of 80,000 to 1,40,000 genes. Almost all (99.9 per cent) nucleotide bases are exactly the same in all people.
4. The functions are unknown for over 50 per cent of the discovered genes.
5. Less than 2 percent of the genome codes for proteins.
6. Repeated sequences make up a very large portion of the human genome.
Q.6. During DNA replication, why is it that the entire molecule does not open in one go? Explain replication fork. What are the two functions that the monomers (d NTPs) play?
Ans. For long DNA molecules, since the two strands of DNA cannot be separated in its entire length (due to very high energy requirement), the replication occur within a small opening of the DNA helix, referred to as replication fork.
Q.7. Retroviruses do not follow central Dogma. Comment.
Ans. Genetic material of retrovirus is RNA. At the time of synthesis of protein, RNA is ‘reverse transcribed’ to its complementary DNA first, which is opposite to the central dogma. Hence, retrovirus are not known to follow central dogma.
Q.8. In an experiment, DNA is treated with a compound which tends to place itself amongst the stacks of nitrogenous base pairs. As a result of this, the distance between two consecutive base increases. from 0.34 nm to 0.44 nm calculate the length of DNA double helix (which has 2×109 bp) in the presence of saturating amount of this compound.
Ans. 2 x 109 x 0.44 x 10-9/bp = 0.88 m.
Q.9. What would happen if histones were to be mutated and made rich in acidic amino acids such as aspartic acid and glutamic acid in place of basic amino acids such as lysine and arginine?
Ans. If histone proteins were rich in acidic amino acids instead of basic amino acids then they may not have any role in DNA packaging in eukaryotes as DNA is also negatively charged molecule. The packaging of DNA around the nucleosome would not happen. Consequently, the chromatin fibre would not be formed.
Q.10. Recall the experiments done by Frederick Griffith, Avery, MacLeod and McCarty, where DNA was speculated to be the genetic material. If RNA, instead of DNA was the genetic material, would the heat killed strain of Pneumococcus have transformed the R-strain into virulent strain? Explain.
Ans. RNA is more labile and prone to degradation (owing to the presence of 2′ OH group in its ribose). Hence heat-killed S-strain may not have retained its ability to transform the R-strain into virulent form if RNA was its genetic material.
Q.11. You are repeating the Hershey-Chase experiment and are provided with two isotopes: 32P and 15N (in place of 35S in the original experiment). How do you expect your results to be different?
Ans. Use of 15N will be inappropriate because method of detection of 35P and 15N is different (32P being a radioactive isotope while 15N is not radioactive but is the heavier isotope of Nitrogen). Even if 15N was radioactive then its presence would have been detected both inside the cell (15N incorporated as nitrogenous base in DNA) as well as in the supernatant because 15N would also get incorporated in amino group of amino acids in proteins). Hence the use of 15N would not give any conclusive results.
Q.12. There is only one possible sequence of amino acids when deduced from a given nucleotides. But multiple nucleotides sequence can be deduced from a single amino acid sequence. Explain this phenomena.
Ans. Some amino acids are coded by more than one codon (known as degeneracy of codons), hence on deducing a nucleotide sequence from an amino acid sequence, multiple nucleotide sequence will be obtained.
For example, lie has three codous: AUU, AUC AUA hence a dipeptide. Met-Ile can have the following nucleotide sequence:
(i) AUG-AUU
(ii) AUG-AUC
(iii) AUG-AUA – And if, we deduce amino acid sequence the above nucleotide sequences, all the three will code for Met-IIe
Q.13. A single base mutation in a gene may not ‘always’ result in loss or gain of function. Do you think the statement is correct? Defend your answer.
Ans. The statement is correct. Because of degeneracy of codons, mutations at third base of codon, usually does not result into any change in phenotype. This is called silent mutations.
Q.14. A low level of expression of lac operon occurs at all the time. Can you explain the logic behind this phenomena.
Ans. In the complete absence of expression of lac operon, permease will not be synthesised which is essential for transport of lactose from medium into the cells. And if lactose cannot be transported into the cell, then it cannot act as inducers, hence cannot relieve the lac operon from its repressed state.
Q.15. How has the sequencing of human genome opened new windows for treatment of various genetic disorders. Discuss amongst your classmates.
Ans. Knowledge about the effects of DNA variations among individuals can lead to revolutionary new ways to diagnose, treat and someday prevent the thousands of disorders that affect human beings.
• Besides providing clues to understanding human biology, learning about non-human organisms DNA sequences can lead to an understanding of their natural capabilities that can be applied toward solving challenges in health care, agriculture, energy production, environmental remediation. Deriving meaningful knowledge from the DNA sequences will define research through the coming decades leading to our understanding of biological systems. .
• This enormous task will require the expertise and creativity of tens of thousands of scientists from varied disciplines in both the public and private sectors worldwide. One of the greatest impacts of having the HG sequence may well be enabling a radically new approach to biological research. In the past, researchers studied one or a few genes at a time. With whole-genome sequences and new high-throughput technologies, we can approach questions systematically and on a much broader scale.
• They can study all the genes in a genome, for example, all the transcripts in a particular tissue or organ or tumor, or how tens of thousands of genes and proteins work together in interconnected networks to orchestrate the chemistry of life.
Q.16. The total number of genes in humans is far less (< 25,000) than the previous estimate (upto 1,40,000 gene). Comment.
Ans. Repeated sequences make up very large portion of the human genome. Repetitive sequences are stretches of DNA sequences that are repeated many times, sometimes hundred to thousand times. They are thought to have no direct coding functions, but they shed light on chromosome structure, dynamics and evolution. Less than 2 per cent of the genome codes for proteins.
Q.17. Now, sequencing of total genomes getting is getting less expensive day by the day. Soon it may be affordable for a common man to get his genome sequenced. What in your opinion could be the advantage and disadvantage of this development?
Ans. Advantages:
1.Knowledge about the effects of DNA variations among individuals can lead to revolutionary new ways to diagnose, treat and some day prevent the thousands of disorders that affect human beings.
2. Besides providing clues to understanding human biology, learning about non-human organisms DNA sequences can lead to an understanding of their natural capabilities that can be applied toward solving challenges in health care, agriculture, energy production, environmental remediation.
3. Deriving meaningful knowledge from the DNA sequences will define research through the coming decades leading to our understanding of biological systems.
4. They can study all the genes in a genome, for example, all the transcripts in a particular tissue or organ or tumor, or how tens of thousands of genes and proteins work together in interconnected networks to orchestrate the chemistry of life.
Disadvantages:
1. It creates the problem of patenting of genes.
2. Persons becomes more careless.
Q.18. Would it be appropriate to use DNA probes such as VNTR in DNA finger printing of a bacteriaphage?
Ans. Bacteriaphage does not have repetitive sequences such as VNTRs in its genome as its genome is very small and have all the coding sequence. DNA finger printing is not done for phages.
Q.19. During in vitro synthesis of DNA, a researcher used 2’, 3’ – dideoxy cytidine triphosphate as raw nucleotide in place of 2’-deoxy cytidine. What would be the consequence?
Ans. Further polymerisation would not occur, as the 3′-OH on sugar is not there to add a new nucleotide for forming ester bond.
Q.20. What background information did Watson and Crick have made available for developing a model of DNA? What was their contribution?
Ans. Watson and Crick had the following information which helped them to develop a model of DNA.
(i) Chargaffs’ law suggesting A = T and C = G.
(ii) Wilkins and Rosalind Franklin’s work on DNA crystal’s X-ray diffraction studies about DNA’s physical structure.
(iii) Watson and crick proposed
a. How complementary bases may pair
b. Semi conservative replication and ‘
c. Mutation through tautomerism
Q.21. What are the functions of (i) methylated guanasine cap, (ii) poly-A “tail” in a mature on RNA?
Ans. Methylated Guanine cap helps in binding of mRNA to smaller ribosomal sub-unit during initiation of translation. Poly-A tail provides longevity to mRNA’s life. Tail length and longevity of mRNA are positively correlated.
Q.22. Do you think that the alternate splicing of exons may enable a structural gene to code for several isoproteins from one and the same gene? If yes, how? If not, why so?
Ans. Functional mRN A of structural genes need not always include all of its exons. This alternate splicing of exons is sex-specific, tissue-specific and even developmental stage-specific. By such alternate splicing of exons, a single gene may encode for several isoproteins and/or proteins of similar class. In absence of such a kind of splicing, there should have been new genes for every protein/isoprotein. Such an extravagancy has been avoided in natural phenomena by way of alternative splicing.
Q.23. Comment on the utility of variability in number of tandem repeats during DNA finger printing.
Ans. Tandemness in repeats provides many copies of the sequence for fingerprinting, and variability in nitrogen base sequence in them. Being individual-specific, this proves to be useful in the process of DNA fingerprinting.
LONG ANSWER TYPE QUESTIONS
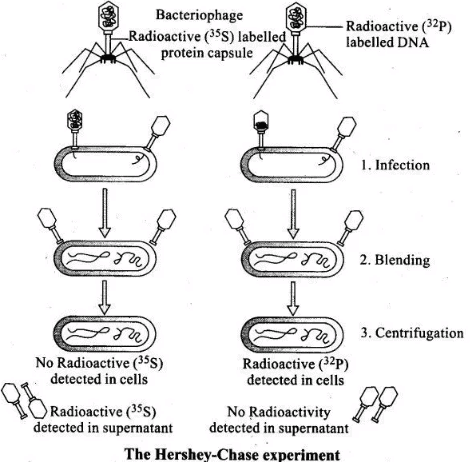
Q.1. Give an account of Hershey and Chase experiment. What did it conclusively prove? If both DNA and proteins contained phosphorus and sulphur do you think the result would have been the same?
Ans. The unequivocal proof that DNA is the genetic material came from the experiments of Alfred Hershey and Martha Chase (1952). They worked with viruses that infect bacteria called bacteriophages. The bacteriophage attaches to the bacteria and its genetic material then enters the bacterial cell. The bacterial cell treats the viral genetic material as if it was its own and subsequently manufacture more virus particles.
• Hershey and Chase worked to discover whether it was protein or DNA from the viruses that entered the bacteria. They grew some viruses on a medium that contained radioactive phosphorus and some others on medium that contained radioactive sulfur. Viruses grown in the presence of radioactive phosphorus contained radioactive DNA but not radioactive protein because DNA contains phosphorus but protein does not
• Similarly, viruses grown on radioactive sulfur contained radioactive protein but not radioactive DNA because DNA does not contain sulfur. Radioactive phages were allowed to attach to E. coli bacteria. Then, as the infection proceeded, the viral coats were removed from the bacteria by agitating them in a blender.
• Radioactive phages were allowed to attach to E. Coli bacteria. Then, as the infection proceeded, the viral coats were removed from the bacteria by agitating them in a blender. The virus particles were separated from the bacteria by spinning them in a centrifuge. Bacteria which was infected with viruses that had radioactive DNA were radioactive, indicating that DNA was the material that passed from the virus to the bacteria.
• Bacteria that were infected with viruses that had radioactive proteins were not radioactive. This indicates that proteins did not enter the bacteria from the viruses. DNA is therefore the genetic material that is passed from virus to bacteria.
Q.2. During the course of evolution why DNA was chosen over RNA as genetic material? Give reasons by first discussing the desired criteria in a molecule that can act as genetic material and in the light of biochemical differences between DNA and RNA.
Ans. A molecule that can act as a genetic material must fulfill the following criteria:
(i) It should be able to generate its replica (Replication):
• Because of rule of base pairing and complementarity, both the nucleic acids (DNA and RNA) have the ability to direct their duplications. The other molecules in the living system, such as proteins fail to fulfill first criteria itself.
(ii) It should chemically and structurally be stable.
• The genetic material should be stable enough not to change with different stages of life cycle, age or with change in physiology of the organism. Stability as one of the properties of genetic material was very evident in Griffith’s ‘transforming principle’ itself that heat, which killed the bacteria,- at least did not destroy some of the properties of genetic material. This now can easily be explained in light of the DNA that the two strands being complementary if separated by heating come together, when appropriate conditions are provided.
• Further, 2′-OR group present at every nucleotide in RNA is a reactive group and makes RNA labile and easily degradable. RNA is also now known to be catalytic, hence reactive. Therefore, DNA chemically is less reactive and structurally more stable when compared to RNA. Therefore, among the two nucleic acids, the DNA is a better genetic material. In fact, the presence of thymine at the place of uracil also confers additional stability to DNA.
(iii) It should provide the scope for slow changes (mutation) that are required
for evolution.
• Both DNA and RNA are able to mutate. In fact, RNA being unstable, mutate at a faster rate. Consequently, viruses having RNA genome and having shorter life span mutate and evolve faster.
(iv) It should be able to express itself in the form of ‘Mendelian Characters’.
• RNA can directly code for the synthesis of proteins, hence can easily express the characters. DNA, however, is dependent on RNA for synthesis of proteins. The protein synthesising machinery has evolved around RNA. The above discussion indicate that both RNA and DNA can function as genetic material, but DNA being more stable is preferred for storage of genetic information. For the transmission of genetic information, RNA is better.
• RNA being a catalyst was reactive and hence unstable. Therefore, DNA has evolved from RNA with chemical modifications that make it more stable.
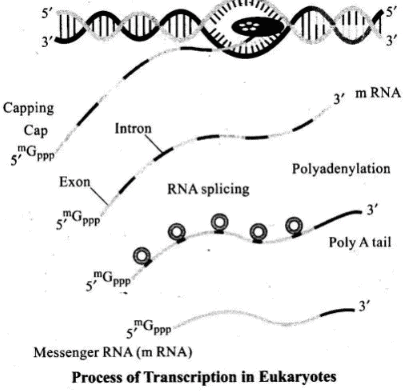
Q.3. Give an account of post transcriptional modifications of a eukaryotic mRNA.
Ans. The primary transcripts (hn-RNA) contain both the exons and the introns and are non-functional. Hence, it is subjected to a process called splicing where the introns are removed and exons are joined in a defined order. Intron is the portion of gene which is transcribed but not translated. In prokaryotes hnRNA is absent so splicing in not required. hnRNA undergoes additional processing called as capping and tailing.
In capping an unusual nucleotide (methyl guanosine triphosphate) is added to the 5′-end of hnRNA. In tailing, adenylate residues (200-300) are added at 3′-end in a template independent manner. It is the fully processed hnRNA, . now called mRNA, that is transported out of the nucleus for translation.
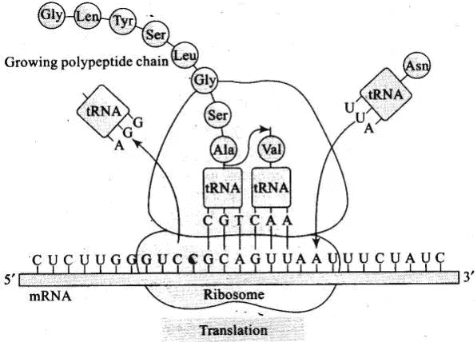
Q.4. Discuss the process of translation in detail.
Ans. Translation refers to the process of polymerisation of amino acids to form a polypeptide. Ribosome is the site of protein synthesis. The amino acids are joined by a bond which is known as a peptide bond. Formation of a peptide bond requires energy. Therefore, in the first phase itself amino acids are activated in the presence of ATP and linked to their cognate tRNA—a process commonly called as activajjon or charging of tRNA or aminoacylation of tRNA to be more specific.
• If two such charged tRNAs are brought close enough, the formation of peptide bond between them would be favoured energetically. The presence of a catalyst would enhance the rate of peptide bond formation. The cellular factory responsible for synthesising proteins is the ribosome. The ribosome consists of structural RNAs and about 80 different proteins. In its inactive state, it exists as two subunits; a large subunit and a small subunit.
• When the small subunit encounters an mRNA, the process of translation of the mRNA to protein begins. There are two sites in the large subunit, for subsequent amino acids to bind to and thus, be close enough to each other for the formation of a peptide bond. The ribosome also acts as a catalyst (23S rRNA in bacteria is the enzyme- ribozyme) for the formation of peptide bond or polyribonucleotides.
• A translational unit in mRNA is the sequence of RNA that is flanked by the start codon (AUG) and the stop codon and codes for a polypeptide. An mRNA also has some additional sequences that are not translated and are referred as untranslated regions (UTR). The UTRs are present at both 5′- end (before start codon) and at 3′-end (after stop codon). They are required for efficient translation process.
• For initiation, the ribosome binds to the mRNA at the start codon (AUG) that is recognised only by the initiator tRNA. The ribosome proceeds to the elongation phase of protein synthesis. During this stage, complexes composed of an amino acid linked to tRNA, sequentially bind to the appropriate codon in mRNA by forming complementary base pairs with the tRNA anticodon. The ribosome moves from codon to codon along the mRNA.
• Amino acids are added one by one, translated into polypeptide sequences dictated by DNA and represented by mRNA. At the end, a release factor binds to the stop codon, terminating translation and releasing the complete polypeptide from the ribosome.
Q.5. Define an operon. giving an example, explain an Inducible operon.
Ans. Lac Operon
• The elucidation of the lac operon was also a result of a close association between a geneticist, Francois Jacob and a biochemist, Jacque Monod in 1961. They were the first to elucidate a transcriptionally regulated system. In lac operon (here lac refers to lactose), a polycistronic structural gene is regulated by a common promoter and regulatory genes.
• Such arrangement is very common in bacteria and is referred to as operon. To name few such examples, lac operon, trp operon, ara operon, his operon, val operon, etc. Lac operon is a type of inducible operon. In inducible operon, presence of a chemical switch on the operon. The lac operon consists of one regulatory gene (the i gene – here the term i does not refer to inducer, rather it Is derived from the word inhibitor) and three structural genes (z, y and a).
• The z gene codes for beta-galactosidase (p-gal), which is primarily responsible for the hydrolysis of the disaccharide, lactose into its monomeric units, galactose and glucose. The y-gene codes for permease, which increases permeability of the cell to p-galactosides. The a-gene encodes a transacetylase.
• Hence, all the three gene products in lac operon are required for metabolism of lactose. In most other operons as well, the genes present in the operon are needed together to function in the same or related metabolic pathway. Operator gene is switched off in the presence of a repressor. RNA polymerase binds with the promoter gene. Lactose is the substrate for the enzyme beta-galactosidase and it regulates switching on and off of the operon. Hence, it is termed as inducer.
• In the absence of a preferred carbon source such as glucose,; if lactose is provided in the grcfwth medium of the bacteria, the lactose is transported into the cells through the action of permease (Remember, a very low level of expression of lac operon has to be present in the cell all the time, otherwise lactose cannot enter the cells). The lactose then induces the operon in the following manner.
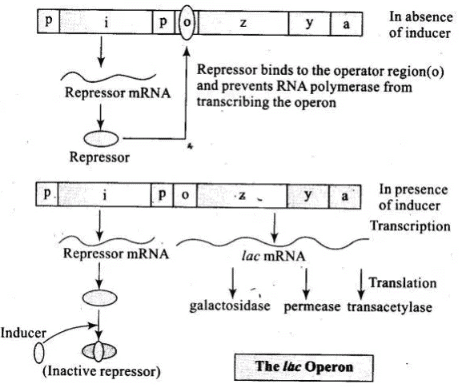
• The repressor of the operon is synthesised (all-the-time constitutively) from the i gene.
Constitutive genes or housekeeping genes: These genes are constantly expressing themselves because their product is required by cell all the time. E.g.: Genes for ATPalse and glycolysis. The repressor protein binds to the operator region of the operon and prevents RNA polymerase from transcribing the operon.
• In the presence of an inducer, such as lactose or allolactose, the repressor is inactivated by interaction with the inducer. This allows RNA polymerase access to the promoter and transcription proceeds. Essentially, regulation of lac operon can also be visualised as regulation of enzyme synthesis by its substrate.
Q.6. ‘There is a paternity dispute for a child’. Which technique can solve the problem. Discuss the principle involved.
Ans. DNA finger printing is used to solve the paternity dispute. DNA fingerprinting involves identifying differences in some specific regions in DNA sequence called as repetitive DNA, because in these sequences, a small stretch of DNA is repeated many times. These repetitive DNA are separated from bulk genomic DNA as different peaks during density gradient centrifugation.
• The bulk DNA forms a major peak and the other small peaks are referred to as satellite DNA. Depending on base composition (A : T rich or G : C rich), length of segment, and number of repetitive units, the satellite DNA
• is classified into many categories, such as micro-satellites, mini-satellites etc. These sequences normally do not code for any proteins, but they form a large portion of human genome.
• These sequence show high degree of polymorphism and form the basis of DNA fingerprinting. Since DNA from every tissue (such as blood, hair- follicle, skin, bone, saliva, sperm etc.), from an individual show the same degree of polymorphism, they become very useful identification tool in forensic applications. Further, as the polymorphisms are inheritable from parents to children, DNA fingerprinting is the basis of paternity testing, in case of disputes.
• The technique of DNA fingerprinting was initially developed by Alec
Jeffreys. Lalji Singh is called father of Indian DNA fingerprinting or DNA profiling or DNA typing. He used a satellite DNA as probe that shows very high degree of polymorphism. It was called as Variable Number of Tandem Repeats (VNTR).
• The technique, as used earlier, involved Southern blot hybridisation using radiolabeled VNTR as a probe. It included
(i) Isolation of DNA,
(ii) Digestion of DNA by restriction endonucleases,
(iii) Separation of DNA fragments by electrophoresis,
(iv) Transferring (blotting) of separated DNA fragments to synthetic membranes, such as nitrocellulose or nylon,
(v) Hybridisation using labelled VNTR probe, and
(vi) Detection of hybridised DNA fragments by autoradiography.
Q.7. Give an account of the methods used in sequencing the human genome.
Ans. Methodologies:
• The methods involved two major approaches. One approach focused on identifying all the genes that are expressed as RNA (referred to as Expressed Sequence Tags (ESTs). The other took the blind approach of simply sequencing the whole set of genome that contained all the coding and non-coding sequence, and later assigning different regions in the sequence with functions (a term referred to as Sequence Annotation).
• For sequencing, the total DNA from a cell is isolated and converted into random fragments of relatively smaller sizes (recall DNA is a very long polymer, and there are technical limitations in sequencing very long pieces of DNA) and cloned in suitable host using specialised vectors. The cloning resulted into amplification of each piece of DNA fragment so that it subsequently could be sequenced with ease.
• The commonly used hosts were bacteria and yeast, and the vectors were called as BAC (bacterial artificial chromosomes), and YAC (yeast artificial chromosomes). The fragments were sequenced using automated DNA sequencers that worked on the principle of a method developed by Frederick Sanger. Sanger is also credited for developing method for determination of amino acid sequences in proteins. These sequences were then arranged based on some overlapping regions present in them.
• This required generation of overlapping fragments for sequencing. Alignment of these sequences was humanly not possible. Therefore, specialised computer based programs were developed. These sequences were subsequently annotated and were assigned to each chromosome. Another challenging task was assigning the genetic and physical maps on the genome. This was’-generated using information on polymorphism of restriction endonuclease recognition sites, and some repetitive DNA sequences known as microsatellites.
Q.8. List the various markers that are used in DNA finger printing.
Ans. Different DNA marker systems such as Restriction Fragment Length Polymorphisms (RFLPs), Random Amplified Polymorphic DNAs (RAPDs), Amplified Fragment Length Polymorphisms (AFLPs), Simple Sequence Repeats (SSRs) which also called as microsatellites, Single Nucleotide Polymorphisms (SNPs) and others have been developed.
Q.9. Replication was allowed to take place in the presence of radioactive deoxynucleotides precursors in E.coli that was a mutant for DNA ligase. Newly synthesised radioactive DNA was purified and strands were separated by denaturation. These were centrifuged using density gradient centrifugation. Which of the following would be a correct result?
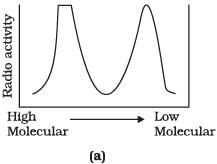
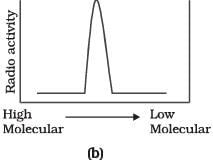
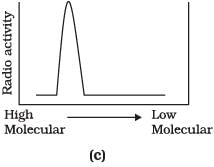
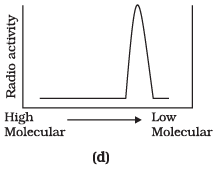
Ans. (d)
|
59 videos|290 docs|168 tests
|
FAQs on NCERT Exemplar: Molecular Basis of Inheritance - 2 - Biology Class 12 - NEET
| 1. What is the molecular basis of inheritance? |  |
| 2. How does DNA replication occur? |  |
| 3. What is the role of transcription in the molecular basis of inheritance? |  |
| 4. How does translation contribute to the molecular basis of inheritance? |  |
| 5. What are the implications of the molecular basis of inheritance in disease development? |  |





















