Molecular basis of infection and disease resistance | Botany Optional for UPSC PDF Download
Introduction
Plants are constantly engaged in an ongoing evolutionary battle against a wide array of microbial and other types of pathogens. These pathogens typically gain access to the interior of plants either by directly penetrating the surfaces of leaves and roots or by entering through wounds and natural openings like leaf stomata. During this invasion process, plant pathogens break down the plant's cell wall by producing and releasing enzymes that degrade it. They then deliver specific pathogen effectors through specialized infection structures and disrupt the normal functions of the host plant (Pajerowska-Mukhtar and Dong, 2009; Tilsner and Oparka, 2010; Horbach et al., 2011).
In response to these threats, plants have evolved intricate defense mechanisms to counteract pathogen invasion. The first line of defense includes the waxy cuticular layers, cell walls, and pre-existing antimicrobial compounds. Once pathogens breach the cell wall, plants activate a two-tiered innate immune system to combat the invaders. The first tier is called PAMP-triggered immunity (PTI), which involves the detection of pathogen- or microbe-associated molecular patterns (PAMPs or MAMPs). The second tier is effector-triggered immunity (ETI), which recognizes effectors produced by pathogens that have evaded PTI (Jones and Dangl, 2006).
In this review, we will explore the genetic and molecular aspects of plant resistance, including the genetic foundations of plant resistance, signaling pathways, the perception of pathogens, and the defense mechanisms employed by plants.
Genetic Basis of Plant Disease Resistance
1. Qualitative and Quantitative Disease Resistance
- Plant disease resistance can be broadly categorized as qualitative and quantitative resistance. Qualitative resistance is controlled by major R genes, while quantitative resistance involves multiple genes with smaller individual effects (Poland et al., 2009).
- R genes typically confer complete resistance to a specific pathogen or pathogen strain and are relatively easy to manipulate for research and crop improvement purposes. However, the effectiveness of resistance mediated by major R genes can be quickly overcome by new, virulent pathogens, posing challenges for plant breeders and farmers.
- Quantitative disease resistance (QDR) is governed by multiple genes, each contributing to partial resistance. QDR exerts lower selection pressure on pathogen variants, making it more durable than R gene-mediated resistance (Parlevliet, 2002). Numerous studies have highlighted the importance of QDR in improving crop disease resistance through traditional or marker-assisted breeding methods.
2. Major Genes for Disease Resistance
Over the past two decades, numerous R genes responsible for qualitative resistance have been identified and cloned from various plant species.
These genes can be categorized into seven classes based on their amino acid motifs and membrane-spanning domains (Gururani et al., 2012):
- NBS-LRR genes, which comprise a nucleotide-binding site (NBS) and a leucine-rich repeat (LRR) domain.
- Extra-cytoplasmic LRR genes with a C-terminal membrane anchor.
- Genes like rice Xa21, which feature both extracellular LRRs and a transmembrane protein kinase.
- Genes such as Arabidopsis RPW8, which contain a membrane protein domain and a coiled-coil domain (CC).
- Genes like Ve1 and Ve2 in tomato, with extracellular LRRs, a PEST domain, and motifs facilitating receptor-mediated endocytosis.
- Genes exemplified by Arabidopsis RRS1-R, which have a C-terminal extension with a nuclear localization signal (NLS) and a WRKY domain.
- Enzymatic genes lacking LRR or NBS domains, such as the maize Hm1 gene, which encodes an enzyme detoxifying a specific fungal toxin.
In addition to dominant resistance genes, recessive resistance genes have also been discovered in plants. These recessive genes, like bs5 and bs6 in peppers or xa34 in rice, control resistance to specific pathogens and are distinct from the dominant R genes. Research suggests that recessive resistance may involve passive mechanisms rather than active defense responses.
For instance, the resistance mediated by a recessive rice xa13 gene can be overcome by a disease susceptibility gene, suggesting that xa13 may act as a susceptibility allele (Iyer-Pascuzzi and McCouch, 2007). Similarly, the pvr2 locus in pepper corresponds to an eukaryotic initiation factor 4E (eIF4E) gene, conferring recessive resistance against potato virus Y (PVY) by disrupting compatibility between the virus's protein and eIF4E in the resistant genotype (Ruffel et al., 2002).
3. Quantitative Loci for Disease Resistance
- Quantitative resistance loci (QRL) may play diverse roles in various biological processes in plants, including the regulation of morphological traits, basal defense, detoxification of pathogen-produced toxins, assistance in defense signal transmission, and interactions with the circadian clock. Some QRL may even represent weaker alleles of R genes or previously unidentified genes.
- The timing of flowering is closely linked to resistance against many pathogens, as susceptibility tends to increase after flowering. This suggests that QRL may be involved in regulating flowering time (Collins et al., 1999). PAMP-triggered immunity, a form of basal defense, provides resistance against a wide range of plant pathogens. QRL in this context may be mutants or alleles of genes associated with basal defense (Dunning et al., 2007). For instance, the level of camalexin, an anti-pathogen chemical in Arabidopsis, is correlated with quantitative disease resistance. Some QRL may be associated with the production of such chemicals.
- Plant defense responses are also regulated by signaling pathways such as the salicylic acid (SA)-dependent, jasmonic acid (JA)-dependent, and ethylene-dependent pathways. Mutations in genes involved in these pathways can affect susceptibility levels, making different alleles of these genes potential QRL (Poland et al., 2009). Recent research has identified novel genes involved in R gene-mediated resistance and their interconnections with the circadian clock, indicating the potential involvement of circadian clock-associated genes in quantitative resistance (Wang et al., 2011b).
- Although only a few QRL associated with quantitative disease resistance have been cloned and characterized to date, they have revealed the diversity of genes and pathways contributing to plant defense. For instance, the pi21 gene in rice, which confers partial resistance to rice blast, encodes a proline-rich protein unrelated to known defense genes (Fukuoka et al., 2009). Similarly, wheat's Yr36 gene, providing resistance to stripe rust, encodes kinase and START lipid-binding domains (Fu et al., 2009). The Lr34 gene in wheat, conferring resistance to multiple pathogens, encodes a putative adenosine triphosphate-binding cassette (ABC) transporter.
Mechanisms of Plant Resistance to Pathogens
1. Biotrophic and Necrotrophic Pathogens
- Plant pathogens can be categorized as biotrophs and necrotrophs, depending on their lifestyles. Biotrophic pathogens obtain nutrients from living host tissue, while necrotrophic pathogens kill host tissue and feed on the dead remains.
- Some pathogens exhibit a hemi-biotrophic lifestyle, behaving as both biotrophs and necrotrophs, depending on conditions or life cycle stages. For example, certain fungi initially act as hemi-biotrophs by infecting the host biotrophically before switching to necrotrophic behavior (Pieterse et al., 2009). The molecular mechanisms of plant defense against biotrophic and necrotrophic pathogens differ significantly.
2. Plant Resistance to Biotrophs
- Plant defense against biotrophic pathogens is primarily driven by gene-to-gene interactions (Glazebrook, 2005). Resistance mediated by R genes typically triggers a hypersensitive response (HR), which restricts biotrophic pathogens like Peronospora parasitica, Pseudomonas syringae, and Erysiphe spp. by limiting their access to water and nutrients (Glazebrook et al., 1997; Aarts et al., 1998; Feys et al., 2001). R gene-mediated resistance also activates a signaling pathway dependent on salicylic acid (SA), leading to the induction of defense effector genes. This SA signaling extends throughout the plant, resulting in systemic acquired resistance (SAR) against subsequent pathogen infections (Glazebrook, 2005). During SAR, the plant deposits callose and lignin in cell walls and acquires the ability to initiate HR rapidly.
- Various Arabidopsis mutants with defects in defense-related signaling pathways support the role of SA signaling in resistance to biotrophic pathogens. Mutations in EDS1 and PAD4, important components of SA signaling, weaken resistance to some P. parasitica isolates. The NPR1 gene, a master regulator of SA signaling, is crucial for plant resistance, and mutants lacking NPR1 are more susceptible to various pathogens (Cao et al., 1994; Bi et al., 2011). Mutations affecting SA synthesis, such as eds5 and sid2, also increase susceptibility to pathogens like P. syringae (Nawrath and Metraux, 1999). Additionally, two plant-specific DNA-binding proteins, SAR Deficient 1 (SARD1) and CBP60g, regulate SA synthesis and play key roles in basal resistance and SAR (Zhang et al., 2010c).
3. Plant Resistance to Necrotrophs
- While gene-for-gene resistance is not commonly associated with resistance to necrotrophic pathogens, some plant pattern recognition receptors (PRRs), such as receptor-like protein kinases (RLKs), are implicated in recognizing necrotrophic fungi (Llorente et al., 2005; Berrocal-Lobo and Molina, 2008). For example, the BIK1 (Botrytis-induced kinase 1) gene encodes a putative RLK involved in early defense against Botrytis cinerea and Alternaria brassicicola (Veronese et al., 2006).
- Host-selective toxins (HSTs) are potent weapons of necrotrophic fungi, causing necrotic lesions upon infection. For instance, the Ptr ToxA toxin from Pyrenophora tritici-repentis induces tan necrosis (Ciuffetti and Tuori, 1999). Phytoalexins, like camalexin in Arabidopsis, play important roles in resistance to necrotrophs. Mutations inhibiting camalexin production, such as the pad3 mutation, reduce resistance to necrotrophic fungal pathogens like A. brassicicola (Thomma et al., 1999a; Zhou et al., 1999). Additionally, defensins, small cysteine-rich proteins, exhibit antimicrobial activity against necrotrophic fungi by disrupting fungal membranes and inhibiting hyphal growth (Aerts et al., 2007).
- Signaling pathways involving jasmonic acid (JA) and ethylene play crucial roles in resistance to necrotrophic pathogens. Mutants with impaired JA signaling, like coi1 mutants, are more susceptible to pathogens such as B. cinerea (Thomma et al., 1998). Similarly, ethylene-insensitive mutants like ein2-1 exhibit increased susceptibility to necrotrophic fungi (Thomma et al., 1999a). These signaling pathways are vital in restricting necrotrophic pathogens, and their activation is associated with defense responses against these pathogens.
- Interestingly, high levels of hypersensitive response (HR) associated with resistance to biotrophs can make plants more susceptible to necrotrophic pathogens. For example, HR does not protect Arabidopsis against B. cinerea infection, a necrotrophic pathogen that infects over 200 plant species (Kliebenstein and Rowe, 2008).
4. RNA Silencing in Plant Resistance
- One of the primary mechanisms plants use to defend against viral infections is RNA silencing (Dinesh-Kumar, 2009). RNA silencing is triggered by double-stranded RNA (dsRNA), which leads to the degradation of RNA sequences homologous to the dsRNA and the silencing of the corresponding genes (Meister and Tuschl, 2004).
- Many plant viruses are RNA viruses, and their replication generates dsRNA that triggers RNA silencing (Waterhouse et al., 1998; Dalmay et al., 2000). Viral effectors can suppress RNA silencing, and various suppressors have been identified in plant viruses (Dinesh-Kumar, 2009). This suppression interferes with the plant's silencing machinery, allowing viruses to evade host defenses.
A two-tiered innate immune system in plants
In plants, there exists a two-tiered innate immune system due to the absence of mobile defender cells and a somatic adaptive immune system. Instead, plants have evolved an innate immune system that effectively detects potential microbial threats and responds to their invasion. This system consists of two tiers: Pattern-Triggered Immunity (PTI) and Effector-Triggered Immunity (ETI). Successful pathogens have developed effectors to suppress PTI, while plants can detect these effectors using Resistance (R) proteins, leading to the activation of ETI. The ongoing co-evolution between plants and pathogens has resulted in some pathogens evolving effectors that can counteract ETI.
1. PTI
- PTI is the first tier of the plant's resistance system, and it is initiated by the recognition of Pathogen-Associated Molecular Patterns (PAMPs) or Microbe-Associated Molecular Patterns (MAMPs) by Pattern Recognition Receptors (PRRs) on the plant cell surface. For example, the receptor FLS2 recognizes the bacterial flagellin component called flg22.
- This recognition triggers a signaling cascade that includes the activation of Mitogen-Activated Protein Kinases (MAPKs) and transcription factors, ultimately leading to immune responses. PTI also involves processes such as the production of reactive oxygen species (ROS), callose deposition, and hormonal changes, including the induction of salicylic acid (SA), jasmonic acid (JA), and ethylene-dependent pathways.
2. ETI
- ETI is the second tier of the plant immune system, where plants have evolved Resistance (R) genes to detect specific pathogen effectors and initiate defense responses. These R genes recognize Avr (Avirulence) genes in pathogens and trigger immunity when the pathogen contains the matching Avr gene.
- Effectors from pathogens can be delivered into plant cells using Type III Secretion Systems (TTSS). For instance, the Pto gene in tomato encodes a protein kinase that activates ETI in response to effectors delivered by Pseudomonas syringae. The interplay between R genes and effectors can switch between compatible (pathogen wins) and incompatible (plant wins) reactions over time.
3. Common Signaling Pathways
- PTI and ETI share several common signaling pathways, including transcriptional reprogramming, programmed cell death (often seen as the Hypersensitive Response or HR), and hormonal changes. However, there are differences in how these pathways are utilized in PTI and ETI.
- For example, the duration of MAPK activity can differentiate between PTI and ETI responses. Additionally, both PTI and ETI can lead to the generation of reactive oxygen species (ROS) as signaling molecules, but their roles may differ. The activation of hormone pathways such as SA, JA, and ethylene can occur in both PTI and ETI, but their outcomes may vary.
In summary, plants have developed a sophisticated two-tiered innate immune system that allows them to detect and respond to pathogen threats effectively. This system involves the initial recognition of PAMPs or effectors, followed by the activation of PTI or ETI responses, respectively, leading to various defense mechanisms to protect the plant from microbial invaders.
Contrasting models of pathogen recognition by plants
In the realm of plant-pathogen interactions, there are contrasting models that seek to explain how plants recognize and respond to pathogens. One of the foundational concepts in this field is the gene-for-gene complementarity, as defined by Flor in the 1930s. According to this principle, single plant resistance genes and corresponding avirulence genes in pathogens play a crucial role in the recognition process, leading to a defense response known as Hypersensitive Response (HR). In simple terms, if both the plant and pathogen have compatible genes, recognition and resistance occur, whereas if either lacks the appropriate gene, the plant becomes susceptible to infection.
1. The Elicitor-Suppressor and Elicitor-Receptor Models
- Two primary models attempt to elucidate the molecular mechanisms behind this gene-for-gene recognition. In the elicitor-suppressor model, pathogens produce general elicitors that activate defense responses in plants until a specific suppressor, produced by a particular pathogen race, counteracts this activation.
- This model posits that many pathogens have substances (elicitors) that nonspecifically trigger plant defenses, but specific suppressors are required to evade these defenses. In contrast, the elicitor-receptor model suggests that proteins or metabolites produced by pathogens, particularly those encoded by avirulence genes (Avr genes), are recognized by specific receptors encoded by plant resistance genes. This specific recognition leads to a resistance response.
- Both models propose that plant Resistance (R) proteins interact with pathogen Avr proteins, but direct molecular evidence for these interactions has been limited, with only a few direct R-Avr interactions demonstrated.
2. The Guard Model
- Recent research supports the "guard model," which suggests that R proteins serve as guards for specific host proteins targeted by pathogen effectors during infection. This model proposes that R proteins indirectly perceive multiple effectors and that relatively few R genes can provide resistance against a wide range of pathogens.
- The guarded effector target, or "guardee," is essential for the effector's virulence function in the absence of the cognate R protein. Experimental examples of this model include the proteins RIN4 and PBS1 in Arabidopsis and RCR3 and Pto in tomato.
3. The Decoy Model
- Another emerging model is the "decoy model," which posits that some host proteins targeted by effectors act as decoys to detect these effectors via R proteins. This model is based on evolutionary dynamics, suggesting that guardees are subjected to opposing natural selection forces based on the presence or absence of polymorphic R genes in plant populations.
- In the presence of functional R genes, guardees are expected to interact more effectively with effectors to enhance pathogen recognition. Conversely, in the absence of R genes, guardees may evolve to decrease their binding affinity with effectors, creating a "decoy" protein specialized in effector perception by R proteins.
- While these models provide insights into plant-pathogen interactions, the complexity of these interactions suggests that different models may apply to specific scenarios. Further experimental evidence is required to distinguish between these models, potentially leading to innovative strategies for manipulating plant innate immunity to enhance pathogen resistance.
Application of major R genes and QRLs
1. Utilization of Major R Genes
- R genes have been extensively employed in crop improvement, but there are inherent risks due to their potential for transient effectiveness and the pathogen's ability to evolve new races. For instance, the powdery mildew outbreak in wheat caused by Blumeria graminis (Bgt) overcame several major resistance genes used in Chinese wheat breeding programs.
- While deploying multiple R genes can provide more durable resistance, the need for approaches that offer long-term effectiveness and specificity remains in plant resistance breeding.
2. Utilization of QRLs
- Quantitative Resistance Loci (QRLs) have been identified in various crop plants, but their practical use in breeding programs has been limited. However, QRLs offer promising applications in crop improvement compared to qualitative resistance due to their broad-spectrum and long-lasting resistance. Genes with minor-to-intermediate additive effects, such as those conferring resistance to rust diseases in wheat and leaf rust in barley, have shown long-lasting resistance effects.
- For example, Fhb1 is a major QRL responsible for resistance to Fusarium head blight (FHB) in wheat. In barley, six QRLs for leaf rust resistance were detected, and some of them were confirmed using marker-assisted backcrossing. In maize, a major QTL called qHSR1 provides resistance to head smut, and this QTL has been successfully introduced into susceptible maize inbred lines via marker-assisted backcrossing, resulting in enhanced resistance without affecting other important agronomic traits. These examples illustrate the potential of QRLs to provide effective and sustainable resistance in crop breeding efforts.
Perspectives
- Understanding the fundamental mechanisms of plant disease resistance is crucial for sustainable agriculture and human health. In recent decades, there have been significant advancements in the field of plant-microbe interactions. These include experimental demonstrations of the role of Pattern Recognition Receptors (PRRs) in plant disease resistance and the discovery that many pathogen virulence factors suppress PRR signaling and associated immune responses known as Pattern-Triggered Immunity (PTI).
- However, it's important to note that research on plant resistance has been somewhat limited, with a heavy focus on a few specific plant-pathogen interactions, mainly involving Arabidopsis and bacterial pathogens. The vast diversity of interactions between plants and microbes suggests that many novel mechanisms remain undiscovered. As we continue to study various plant-pathogen systems, advances in genome sequencing and the isolation of Resistance (R) genes will greatly aid our understanding of molecular plant-microbe interactions.
- In recent years, detailed models of plant-pathogen interactions have emerged, encompassing the processes of recognition, evasion, and defense. However, it's likely that the molecular basis of plant resistance involves an even broader range of mechanisms. Components of signal transduction systems, antimicrobial compounds like phytoalexins, and other unknown factors are also expected to play significant roles in plant defense responses, and these aspects require further characterization.
- Cloning additional resistance genes and Quantitative Trait Loci (QTLs) associated with plant resistance will provide insights into how they contribute to plant defenses. This knowledge will enable more efficient and effective utilization of these genes in crop improvement and protection strategies.
Figure Legends
Figure 1: Molecular Mechanisms of Plant Resistance to Pathogens
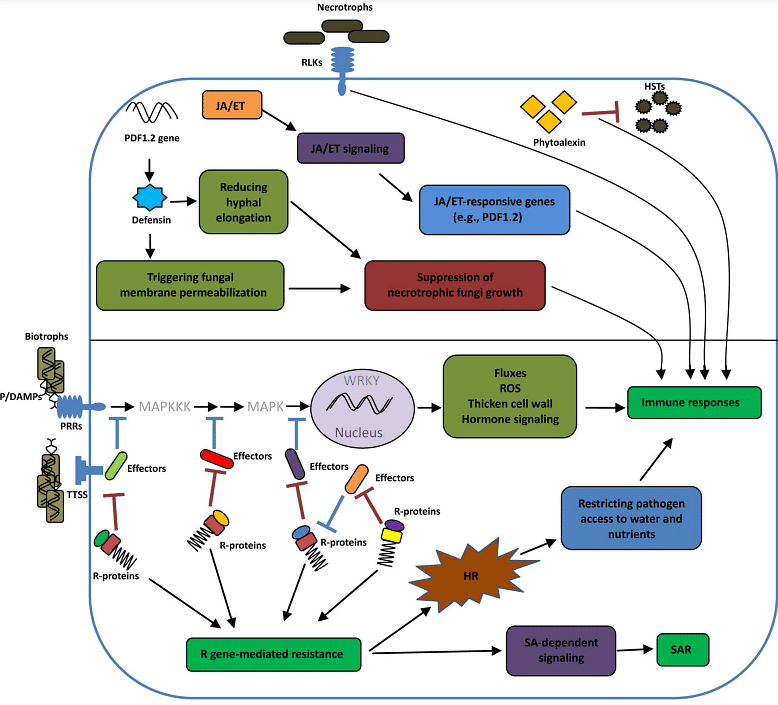
This figure illustrates the molecular basis of plant resistance to both necrotrophic and biotrophic pathogens. In the upper part of the diagram, the defense response to necrotrophic pathogens is depicted. It is conferred by receptor-like kinases (RLKs), defensins, phytoalexins, and signaling pathways involving jasmonic acid (JA) and ethylene (ET). In the lower part of the diagram, the two-tiered immune system of plant resistance to biotrophic pathogens is shown. The first tier, known as Pattern-Triggered Immunity (PTI), is initiated upon the recognition of Pathogen/Damage-Associated Molecular Patterns (P/DAMPs) by membrane-anchored Pattern Recognition Receptors (PRRs). This recognition leads to the activation of the Mitogen-Activated Protein Kinase (MAPK) cascade and downstream transcription factors, ultimately resulting in immune responses. The second tier of defense, known as Effector-Triggered Immunity (ETI), is activated when pathogen effectors interact with specific Resistance (R) proteins. This interaction can oscillate between compatible (susceptibility) and incompatible (resistance) reactions over time. ETI-mediated resistance often leads to hypersensitive response (HR) and also activates signaling pathways involving salicylic acid (SA), leading to systemic acquired resistance (SAR). The figure also includes abbreviations for various components and processes involved.
Figure 2: Contrasting Models of Plant-Pathogen Interaction
 This figure provides a comparison between two models explaining the molecular basis of plant-pathogen interactions.
This figure provides a comparison between two models explaining the molecular basis of plant-pathogen interactions.
A: The Elicitor-Suppressor Model - In this model, pathogens initially release elicitors that trigger plant defense reactions (resistance). However, the resistance can be overcome when a specific suppressor is produced by a particular pathogen race, leading to a failure of the defense reaction (susceptibility).
B: The Elicitor-Receptor Model - In contrast, this model posits that proteins encoded by avirulence genes (Avr genes) in pathogens are recognized by specific plant receptors. This recognition then triggers a robust resistance response. If the receptor in the plant does not match the avirulence protein, susceptibility is likely to occur.
Figure 3: Contrasting Models of Effector Recognition in Plant Resistance
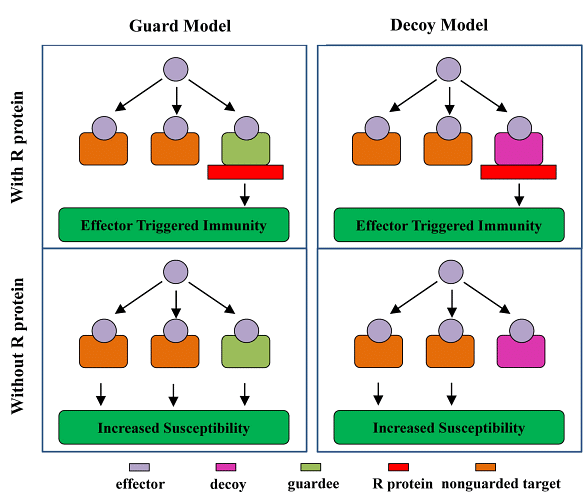 This figure presents a comparison between two models explaining how plants recognize pathogen effectors.
This figure presents a comparison between two models explaining how plants recognize pathogen effectors.
A: The Guard Model - In this model, multiple effectors can be perceived by a single Resistance (R) protein. These R proteins act as guards for specific host proteins (guardees) that are targeted by pathogen effectors during infection. The guard model suggests that a few R genes can provide resistance against a broad spectrum of pathogens.
B: The Decoy Model - According to this model, the guardee proteins are in an evolutionarily unstable situation known as "decoy." In the presence of functional R genes, natural selection favors guardees that interact effectively with effectors to enhance pathogen detection. In contrast, in the absence of R genes, guardees may reduce their binding affinity with effectors, evading detection and modification by the effector. This process can lead to the evolution of host "decoy" proteins specialized in effector perception.
|
179 videos|140 docs
|

|
Explore Courses for UPSC exam
|

|

















