Methods of Transfer of Genes | Botany Optional for UPSC PDF Download
| Table of contents |

|
| Introduction |

|
| Methods of Gene Transfer |

|
| Methods for gene transfer in plants |

|
| Electroporation |

|
| Microinjection |

|
| Selection of Transformants |

|
Introduction
Fertility barriers are surmounted to artificially introduce foreign genes into crops, a crucial process within genetic engineering, commonly referred to as genetic transformation.
Methods of Gene Transfer
- Plant transformation techniques typically encompass the following steps:
- Introducing a DNA segment into totipotent cells,
- Integrating this DNA into the host cell's genome, and
- Subsequently regenerating whole plants from these transformed cells.
Therefore, effective plant transformation methods require a means to efficiently introduce DNA into cells and then regenerate these transformed cells or tissues into complete plants.
- The DNA segment introduced during this process includes the target gene and a cassette containing supplementary genetic material. This supplementary genetic material consists of:
- A promoter that dictates when and where the introduced gene is expressed,
- A terminator that marks the end of transcription, and
- A marker gene that enables the selection of plants carrying the introduced gene.
- Nearly 150 plant species have successfully incorporated and consistently expressed desirable traits through genetic transformation. These methods for achieving genetic transformation in plants, i.e., the delivery of foreign DNA into the host plant, can be categorized into two main groups:
- Indirect methods, where a vector is required to insert foreign DNA into the host genome, and
- Direct methods, which do not rely on a vector and instead involve the direct insertion of DNA into the host genome.
Methods for gene transfer in plants
In-direct Methods
Agrobacterium-mediated genetic transformation
- Indirect methods of genetic transformation, such as Agrobacterium-mediated genetic transformation, utilize bacteria as a carrier to introduce the gene construct into the target cell. This approach involves the use of Agrobacterium, a gram-negative soil bacterium known for causing crown gall disease in various plants, in plant transformation experiments. The most extensively studied species within this genus is Agrobacterium tumefaciens, which serves as an efficient delivery system for genetically transforming plants.
- These bacteria contain a sizable plasmid known as the Ti plasmid (tumor-inducing plasmid), housing genes responsible for inducing tumors (T-DNA) and other genes involved in integrating T-DNA into the host genome. When plants are wounded, they release a sap rich in phenolic compounds that act as chemical attractants for Agrobacteria and stimulate the expression of virulence genes. This leads to the infection of the plant by Agrobacterium, the insertion of the T-DNA region into a random location in the host genome, and the subsequent proliferation of plant cells, resulting in the formation of crown gall growth.
- Another frequently employed species is Agrobacterium rhizogenes, which induces the development of hairy roots in plants. It carries the Ri plasmid (root-inducing plasmid). The Agrobacterium genus exhibits a broad host range and can infect numerous dicotyledonous plants, as well as some monocots.
 Crown gall disease caused by Agrobacterium on rose stem
Crown gall disease caused by Agrobacterium on rose stem
 Ti plasmid with T-DNA region
Ti plasmid with T-DNA region
Structure of Ti Plasmid
- In genetic transformation experiments, a vector serves as the carrier responsible for transporting the gene of interest, along with the promoter, terminator, and selectable marker genes, into the host plant's DNA. The virulence of Agrobacterium is conferred by the Ti plasmid, which contains genes responsible for inducing tumors, integrating T-DNA, and synthesizing plant hormones and opines.
- Origin or Replication Region: This portion of the Ti plasmid is responsible for its replication independently of the bacterium's cell.
- Virulence Region: Within this region are genes known as vir genes, whose products facilitate the processing and transfer of T-DNA from the bacterium to plant cells. These genes are activated in response to specific phenolic compounds like acetosyringone, which are released by plants when they are wounded.
- T-DNA Region: This region of the Ti plasmid contains genes responsible for tumor induction. It is flanked by 25-base pair direct repeat sequences on both ends, known as the Left border (LB) and Right border (RB). Among the various genes in this region are iaaM and iaaH, which are responsible for synthesizing indole acetic acid (an auxin), the ipt gene, which synthesizes an enzyme called isopentenyl adenine (a cytokinin), and the tml gene, another gene involved in tumor formation.
- Opine biosynthesis genes in this region lead to the production of opines, ultimately resulting in the overgrowth of tissue within plant cells, leading to tumor formation.

- Region of Opine Catabolism: This section of the plasmid contains several other genes involved in the metabolism of opines. Importantly, this region is not transferred to plant cells during the infection process.
Use of Ti plasmid in genetic transformation
- In the context of genetic transformation, the Ti plasmid is employed as a vector, with most of its T-DNA region being replaced by the gene of interest, while the left and right border sequences are retained. It's important to note that the T-DNA region is defined not by its specific genetic sequence but by the presence of its borders, which enable its insertion into the host plant's genome.
- The wild-type Ti plasmid from Agrobacterium cannot be directly used as a vector system due to its large size, the presence of genes that cause tumors, and the absence of marker genes and unique restriction sites within the T-DNA region. Consequently, these plasmids are genetically modified to incorporate these necessary elements.
There are two main types of genetically engineered Ti plasmid-based vectors:- Binary Vectors: In this system, a pair of plasmids, which are independent of each other, coexist within the same Agrobacterium cell. This vector system is based on the idea that the vir genes do not need to be present in the same plasmid as the T-DNA for its transfer.
- Helper Ti Plasmid: This plasmid contains the vir region required for T-DNA integration.
- A Disarmed Ti Plasmid (Mini Ti Plasmid): This separate plasmid houses the T-DNA region containing the gene of interest and the plant selectable marker gene, along with their associated promoter and terminator sequences. Additionally, it contains origins of replication for both Agrobacterium and E. coli.
- Co-Integrate Vectors: These vectors, also known as hybrid Ti plasmids, incorporate both the T-DNA region (containing the gene of interest) and the vir genes within the same plasmid used for transformation.
- Binary Vectors: In this system, a pair of plasmids, which are independent of each other, coexist within the same Agrobacterium cell. This vector system is based on the idea that the vir genes do not need to be present in the same plasmid as the T-DNA for its transfer.
Here are the steps involved in the Agrobacterium-mediated genetic transformation of plants using the 'Wounded explant' method, paraphrased:
- Isolate/amplify the gene of interest from a source organism.
- Create an expression cassette, which includes the gene of interest flanked by promoter and terminator sequences for gene expression, along with marker genes to aid in the selection of transformed plants by tracking the introduced genes in the host plant.
- Insert the expression cassette into the T-DNA region of either a binary vector or a co-integrate vector.
- Carry out the transformation of the expression vector into Agrobacterium.
- Obtain explants from the plant intended for transformation. These explants are wounded and then co-cultured briefly to allow for Agrobacterium infection. This co-culturing is achieved using tissue culture techniques.
- Cultivate the transformed explants in the presence of a bacteriostatic agent to inhibit Agrobacterium growth, and in the presence of a selective antibiotic to prevent the survival of any untransformed explants.
- Use PCR analysis to test the transformed explants and confirm the presence of the desired gene.
- Maintain the transformed explants in vitro until they are ready for transplantation.
- After obtaining transformed plants, isolate their DNA and conduct Southern hybridization to confirm the integration site and determine the copy number of the desired gene.
Direct Methods
Direct methods of genetic transformation do not rely on bacteria as intermediaries for integrating DNA into the host genome. These methods include microprojectile bombardment, electroporation, and microinjection.
Microprojectile/Particle Bombardment (Biolistics)
- Biolistics is a technique where cells are physically introduced to nucleic acids or other biological molecules. A biolistic particle delivery system is a device used in plant transformation, where cells are bombarded with heavy metal particles coated with DNA or RNA. This method was developed by John Stanford in 1984 as a means to introduce DNA into cells through physical means, thus avoiding the limitations associated with the host-range restrictions of Agrobacterium. While Agrobacterium-mediated genetic transformation works well for dicotyledonous plants, it has lower efficiency for monocots. Biolistic particle delivery systems offer an effective and versatile approach to transforming a wide range of cell types. This method has successfully produced transgenic organisms in prokaryotes, mammals, and various plant species.
- In this process, the construct containing the gene of interest is coated onto tiny gold or tungsten particles (0.6 – 1 mm in size). Prior to coating, DNA is precipitated with calcium chloride, spermidine, and polyethylene glycol. These coated microparticles are loaded onto a macrocarrier and accelerated to high speeds using pressurized helium gas. Various plant cell suspensions, callus cultures, or tissues can be used as targets for these microparticles. As the microparticles penetrate the plant cell walls and membranes, the coated DNA is released from their surface and becomes incorporated into the plant's genome. Unlike Agrobacterium-mediated transformation, biolistics does not require the use of binary vectors with T-DNA border sequences.
- This method is particularly valuable for monocots, where the efficiency of other transformation techniques is often unsatisfactory. It can be applied to a wide range of tissues, including apical and floral meristems, embryos, seedlings, leaves, cultured cells, and floral tissues.
Several factors must be carefully considered before using particle bombardment, which can be categorized into three main areas:- Physical Parameters: These include the nature, chemical and physical properties of the metal particles used to carry the foreign DNA, the characteristics and preparation of DNA, the binding of DNA to the particles, and the nature of the target tissues.
- Environmental Parameters: Variables such as temperature, photoperiod, and humidity levels in donor plants, explants, and bombarded tissues can affect tissue physiology and impact the receptiveness of the target tissue.
- Biological Parameters: Factors such as the choice and nature of explants, pre- and post-bombardment culture conditions, and osmotic pre- and post-treatment of explants also play a role in the success of the transformation process.
Advantages of Particle Bombardment over Agrobacterium-Mediated DNA Transfer:
- Species Independence: Particle bombardment is not limited by the species and has been successfully applied to a wide range of organisms.
- Overcoming Recalcitrance: It is effective for species that are challenging to transform using other direct methods or are not easily amenable to Agrobacterium-mediated transformation.
- Simplified DNA Requirements: Unlike Agrobacterium-mediated transformation, particle bombardment does not require the inclusion of sequences necessary for T-DNA replication and transfer since it bypasses the complex interaction between the bacterium and plant tissue.
- Transformation of Organelle DNA: This method has been used to achieve the transformation of organelle DNA, including mitochondria and chloroplasts.
- Multigene Introduction: Particle bombardment allows for the introduction of multiple genes into a single plant, enabling the simultaneous modification of several traits.
- Versatile Nucleic Acid Delivery: Particles can be coated with various nucleic acids, including DNA, RNA, siRNA, and large nucleic acid fragments.
Limitations of Particle Bombardment Method:
- Limited Regeneration Capacity: The tissue being bombarded may have limited capacity for regeneration, which can affect the overall success of the transformation.
- Efficiency of Integration: The stable integration of DNA can be less efficient compared to other methods, and the introduced DNA may not always integrate effectively.
- Multiple Copies of the Gene: Particle bombardment can result in the insertion of multiple copies of the gene of interest, leading to potential complications in the plant.
- Integration of Rearranged/Truncated DNA: There is a risk of integrating rearranged or truncated DNA sequences, which may not function as intended.
- Cellular Tissue Damage: The process of bombardment can cause damage to cellular tissues, potentially impacting the health and development of the transformed cells.
- Specialized and Expensive Equipment: Particle bombardment requires specialized and often costly equipment, which may not be readily available in all laboratories.
Electroporation
- Electroporation is a method used for direct gene transfer in which a mixture containing cells and DNA is exposed to very high-voltage electrical pulses (typically between 4000 and 8000 V/cm) for extremely brief durations, often just a few milliseconds. This process results in the formation of temporary pores in the plasma membrane through which DNA can enter the cell and subsequently reach the cell nucleus.
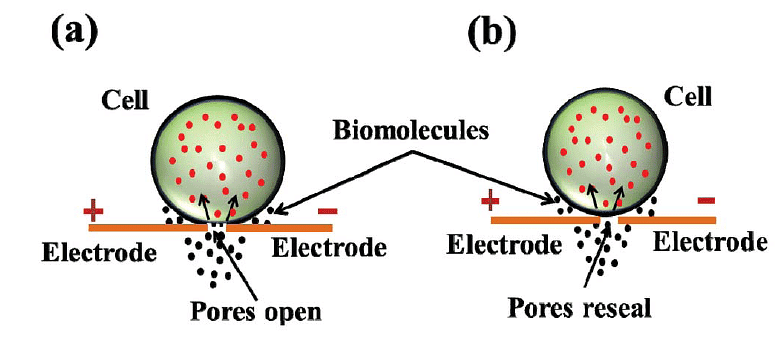 Electroporation (A) Diagram showing formation of transient pores in cell membrane on applying electrical pulse, entry of DNA inside thecell and sealing of pores afterwards.
Electroporation (A) Diagram showing formation of transient pores in cell membrane on applying electrical pulse, entry of DNA inside thecell and sealing of pores afterwards.
- The procedure involves taking a suspension of cells along with plasmid DNA and placing them in an electroporation cuvette positioned between electrodes. When electrical pulses are applied, these brief high-voltage shocks create temporary micropores in the cell membranes. These micropores allow the cells to uptake plasmid DNA, leading to either stable or transient DNA expression within the cells.
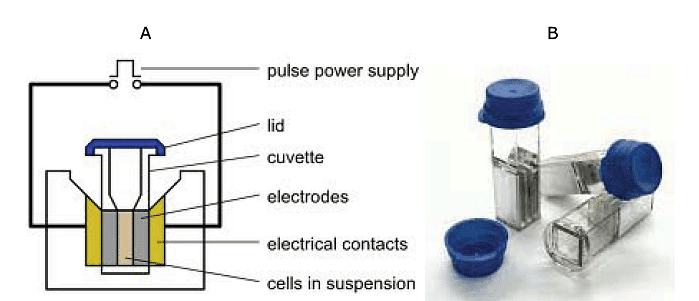 (A) Main components of an electroporator. (B) Cuvettes used for electroporation. These are plastic cuvettes with lid and aluminium electrodes, having a maximum capacity of 400 μ
(A) Main components of an electroporator. (B) Cuvettes used for electroporation. These are plastic cuvettes with lid and aluminium electrodes, having a maximum capacity of 400 μ
- Cells arrested at the metaphase stage of the cell cycle are particularly suitable for electroporation because they lack a nuclear envelope and have a unique permeability of the plasma membrane. Protoplasts, which are plant cells with their cell walls removed, are often used for electroporation in plant cells, as the thick plant cell walls can hinder the movement of DNA. Initially developed for protoplasts, the electroporation method has also yielded successful results with cells and even tissues, allowing for the recovery of regenerated plantlets. In the case of maize, immature zygotic embryos and embryogenic calli have been utilized for electroporation to produce transgenic plants.
- While electroporation allows for the transformation of protoplasts, it is associated with lower transient expression of transgenes compared to organized tissues and has a lower regeneration frequency, especially in monocotyledonous plants. The electric field and chemical substances used to disrupt cell walls can significantly reduce the viability and capacity for division of protoplasts.
- Electroporation is valued as a transformation method for its speed, convenience, simplicity, and cost-effectiveness, and it has a low level of cell toxicity. However, one of its drawbacks is the difficulty in regenerating plants from protoplasts when this specific cell type is used for electroporation.
Microinjection
- Microinjection is a process that involves using a fine glass micropipette to manually inject transgenes at a microscopic or borderline macroscopic level. The transgene can take the form of plasmids, cosmids, phage, YACs (Yeast Artificial Chromosomes), or PCR products, and it may be either circular or linear. Physical linkage of the transgenes is not a requirement for injection.
- In microinjection, DNA is mechanically introduced directly into the nucleus or cytoplasm of target cells using a glass microcapillary injection pipette. Protoplasts (plant cells with their cell walls removed) are immobilized in low-melting agar during the procedure. A microscope is used to work with the cells, and a holding pipette and suction force are employed to keep the cells in place. DNA is then injected directly into the cytoplasm or nucleus of these cells. Subsequently, the injected cells are cultured in vitro and regenerated into plants. Successful examples of plant transformation via microinjection have been demonstrated in crops like rapeseed, tobacco, and various other plant species.
- While microinjection can lead to the generation of stable transformants, it requires a high level of technical expertise and is a time-consuming process. Additionally, microinjection has achieved only limited success in plant transformation due to the presence of thick cell walls in plants and the lack of a widely available single-cell-to-plant regeneration system in most plant species.
- Microinjection is typically performed using one of three types of microscopes: a traditional compound microscope (approximately 200X magnification), an inverted microscope (also around 200X magnification), or a dissecting stereomicroscope (around 40-50X magnification). Under the microscope, the target cell is positioned, and both the cell membrane and nuclear envelope are penetrated with the help of two micromanipulators. One micromanipulator holds the pipette, while the other holds the microcapillary needle.
- There are two main types of microinjection systems: constant flow and pulsed flow:
- Constant Flow System: In this system, the amount of sample injected is determined by the duration for which the needle remains in the cell. It is relatively simple and cost-effective but considered outdated.
- Pulsed Flow System: This system provides greater control over the volume of substance delivered, needle placement and movement, and offers better precision. It results in less damage to the receiving cell. However, the components of this system tend to be expensive.
Chemical-Mediated Gene Transfer
Chemicals can be used to stimulate cells or protoplasts to take up foreign DNA. One of the most commonly used chemicals for this purpose is Polyethylene Glycol (PEG). PEG assists in the precipitation of DNA, which can then be internalized by the cells through the process of endocytosis.
Liposome-Mediated Gene Transfer
In this method, plasmids containing the desired foreign genes are enclosed within small lipid structures called liposomes. These liposomes can then be fused with protoplasts using chemicals like PEG. This allows the transfer of the foreign DNA into the target cells.
Silicon Carbide Method
The silicon carbide method involves the use of organic material fibers, such as silicon carbide, for gene transfer. When these fibers are mixed with plasmid DNA and plant tissue or cells, they aid in the penetration of the foreign DNA into the plant tissue.
Selection of Transformants
In genetic transformation experiments, the uptake of the transgene by cells or organisms is a rare event, occurring in only one in several million to billion cells, depending on the efficiency of the transformation process. Instead of individually examining every single cell or organism, a selective agent can be employed to identify the transformed ones. These selective genes are known as marker genes, and they serve the dual purpose of indicating successful transformation and assessing the success rate of the genetic transformation study.
Marker Gene
A marker gene is introduced into cells along with the transgene to determine if the transgene has been successfully integrated into the host organism's genome. The presence of the marker gene can be visualized or detected.
There are two main types of marker genes:
- Selectable Marker: A selectable marker is a gene that imparts a trait suitable for artificial selection because it protects the organism from a selective agent that would normally be lethal or inhibit its growth. Since only a very small fraction of cells take up the transgene in most genetic transformation experiments, a selective agent is used to eliminate cells without the transgene, leaving behind only the transformed ones.
Selectable marker genes can be categorized based on whether they result in positive or negative selection and whether selection is conditional or non-conditional on the presence of external substrates.- Negative Selectable Marker: Negative selectable marker genes cause the death of untransformed tissue, while transformed cells can survive. Common selective agents include antibiotics and herbicides. When grown in a medium containing the selective agent, non-recombinant cells die due to their lack of resistance. Examples of marker genes include hptII, nptII, and bar, among others.
- Positive Selectable Marker: Positive selectable marker genes promote the growth of transformed tissue without harming untransformed tissue. Initially, positive selectable marker genes were conditional on the use of toxic agents such as antibiotics, herbicides, or drugs. More recently, positive selectable marker genes have been developed that are conditional on non-toxic agents, such as compounds that induce growth and differentiation of transformed tissues. Some of the new-generation positive selectable marker genes do not depend on external substrates but instead modify normal physiological processes involved in plant development. An example is the Pmi gene derived from E. coli, which codes for the phosphomannose isomerase enzyme used as a positive selection marker.
- Screening Marker: A screening marker is a type of marker used to distinguish between cells or organisms that contain a specific gene and those that do not. Unlike selectable markers that provide a selective advantage, screening markers make transformed cells or organisms look different, enabling researchers to manually separate the wanted from the unwanted.
These markers are often used in reporter systems to determine various aspects, including the intracellular localization of a gene product, the efficiency of gene delivery systems, the detection of protein-protein or protein-DNA interactions, and the activity of promoters. One common example of a reporter system is the "blue and white" selection in bacteria, which relies on the insertional inactivation of the LacZ gene that produces beta-galactosidase enzyme. This enzyme converts a colorless substrate into a blue-colored product, resulting in blue colonies. Non-recombinant colonies, which have an intact LacZ gene, remain blue, while recombinants have an insertionally inactivated LacZ gene, resulting in colorless colonies.
There are several types of screening markers commonly used:- GUS Assay: The GUS (β-glucuronidase) assay is a straightforward method for detecting transformed cells without the need for selective agents. The β-glucuronidase enzyme from Escherichia coli is used in this technique. This enzyme can transform colorless or non-fluorescent substrates into colored or fluorescent products, giving transformed cells a distinct phenotype. Histochemical GUS staining uses X-gluc (5-bromo-4-chloro-3-indolyl glucuronide), which produces a blue color when incubated with tissue expressing the GUS gene. For fluorimetric estimation, 4-methylumbelliferyl-beta-D-glucuronide (MUG) is used. This substrate produces a fluorescent product upon hydrolysis, allowing for quantitative analysis. However, a drawback of this technique is that it kills the cells in the process.
- GFP (Green Fluorescent Protein): The 2008 Nobel Prize in Chemistry was awarded for the discovery and development of GFP. GFP is a protein that exhibits bright green fluorescence when exposed to blue to ultraviolet light. Cells containing GFP glow green under UV light and can be visualized using a specialized microscope. GFP has been used as a reporter gene to confirm the expression of a transgene throughout an organism. It can be introduced into organisms through breeding, cell transformation, or viral vector injection. GFP has been expressed in various organisms, including bacteria, fungi, plants, fish, flies, and mammalian cells. Different mutants of GFP have been engineered, leading to color variants such as yellow and red fluorescence, allowing for the tracking of multiple genes simultaneously in an organism. Mutants like EGFP, superfolder GFP, and various color variants have expanded the utility of GFP in research.
Luciferase
- Bioluminescence is the production and emission of light by a living organism, and it is a form of chemiluminescence where a chemical reaction generates light energy. This phenomenon occurs in various organisms, including simple unicellular ones like bacteria and dinoflagellates, as well as more complex organisms like fish and insects. Bioluminescence is often observed in marine animals, especially those living in the depths of the ocean, and it serves various natural purposes, including defense, camouflage, attracting prey or mates, and aiding in feeding and mating.
- Luciferases are enzymes responsible for emitting light in bioluminescence. Many organisms use luciferase-mediated bioluminescence to achieve different objectives, such as attracting prey, finding mates, or deterring predators. For example, firefly luciferase from the firefly species Photinus pyralis emits green light when it catalyzes the oxidation of its chemical substrate, luciferin. When other organisms express the luciferase (LUC) gene and are provided with luciferin, they also emit faint green light.
- The bioluminescence reaction involves two key components: the enzyme (e.g., firefly luciferase) that catalyzes the reaction and the substrate (luciferin). The reaction occurs in two steps:
- Luciferin + ATP ⟶ Luciferyl Adenylate + O2
- Luciferyl Adenylate + O2 ⟶ Oxyluciferin + Light
- Oxyluciferin, formed at the end of this reaction in an electronically excited state, emits a photon of light as it returns to the ground state. Instruments like luminometers or modified optical microscopes are used to detect this emitted light. Luciferase bioluminescence doesn't require external light excitation, resulting in minimal autofluorescence, which minimizes background interference. This bioluminescent glow serves as an assay for LUC (Luciferase gene) expression, acting as a "reporter" to assess the activity of regulatory elements. Since different sources of Luciferase enzymes may exhibit inherent variability in light emission, researchers often use two or more luciferase enzymes in combination for the analysis of multiple genes.
Summary
- Gene transfer involves the introduction of foreign genetic material into a cell, whether artificially or naturally.
- Genetic transformation is the process of introducing foreign genes into crops, a crucial step in genetic engineering.
- Plant transformation typically consists of introducing a DNA segment into plant cells, integrating it into the host cell's genome, and regenerating the transformed cells into whole plants.
- The introduced DNA segment contains the gene of interest flanked by promoter, terminator, and marker genes.
- Genetic transformation methods can be categorized into two main types: direct and indirect.
- Indirect methods use bacteria, such as Agrobacterium, as vectors to introduce gene constructs into target cells. Agrobacterium carries a Ti plasmid with T-DNA for integration into the plant genome.
- Genetically engineered Ti plasmids are used as vectors, with binary and cointegrate vectors being common types.
- Direct methods, on the other hand, do not use bacteria as intermediaries for gene integration. These methods include microprojectile bombardment, electroporation, and microinjection.
- Biolistics is a method that involves bombarding cells with heavy metal particles coated with DNA/RNA, enabling DNA transfer into plant cells.
- Electroporation exposes cells to high-voltage electrical pulses, creating transient pores in the plasma membrane for DNA entry.
- Microinjection manually injects transgenes into cells using fine glass micropipettes.
- Selection of transgenic cells is crucial in transformation experiments, often achieved using selectable marker genes that protect cells from selective agents.
- Screening markers are used to visually distinguish transgenic cells or organisms from non-transformed ones, examples include Gus, GFP, and Luciferase.
|
179 videos|140 docs
|

|
Explore Courses for UPSC exam
|

|

















