Regulation of Gene Expression | Botany Optional for UPSC PDF Download
| Table of contents |

|
| Introduction |

|
| Overview of Regulation of Gene Expression |

|
| Prokaryotic Gene Regulation |

|
| Eukaryotic Gene Regulation |

|
Introduction
- In the human body, virtually all somatic cells share the same DNA, with only a few exceptions like mature red blood cells and certain immune cells that modify their DNA while producing antibodies. Despite having identical DNA, cells in different tissues or organs exhibit distinct structures and functions. This raises the question of how cells can vary so significantly even when they possess the same genetic information.
- The answer lies in the concept of gene expression, which is the process by which cells control which genes are activated and which ones remain dormant. Although each cell contains the same set of genes in its DNA, not all of these genes are active simultaneously. Instead, each cell selectively expresses a specific subset of genes, determining the proteins it produces. Cells in the eye, for instance, express a unique set of proteins distinct from those produced by liver cells. This regulation of gene expression is a highly intricate process involving multiple levels and stages of control, ensuring that the right proteins are synthesized in the correct cells at the appropriate times.
Overview of Regulation of Gene Expression
- Proper cellular function relies on the timely synthesis of necessary proteins. All cells, regardless of their complexity, regulate the synthesis of proteins based on the information stored in their DNA. This process, known as gene expression, involves activating genes to produce mRNA and subsequently proteins. Cells carefully manage when genes are turned on, how much protein is manufactured, and when production should cease due to the protein no longer being required.
- Regulating gene expression offers significant energy and space conservation benefits. It is more energy-efficient to activate genes only when their products are needed. Moreover, limiting the expression of specific genes in each cell conserves space, as DNA must unwind from its tightly coiled structure to facilitate transcription and translation. If every protein were expressed in every cell at all times, cells would have to be excessively large. Gene expression control is a highly intricate process, and any disruptions can have detrimental effects on the cell, potentially leading to the development of various diseases, including cancer.
Prokaryotic versus Eukaryotic Gene Expression
- Prokaryotic organisms, being single-celled and lacking a defined cell nucleus, have their DNA freely floating in the cytoplasm. When a particular protein is needed, the corresponding gene is transcribed into mRNA, which is then immediately translated into protein. Transcription ceases when the protein is no longer required. In prokaryotic cells, regulation primarily occurs at the transcription stage, controlling how much of each protein is expressed.
- In contrast, eukaryotic cells are more complex, featuring intracellular organelles. In eukaryotic cells, DNA resides within the nucleus and is transcribed into mRNA. The newly synthesized mRNA undergoes modifications and exits the nucleus, entering the cytoplasm where ribosomes facilitate translation into protein. Transcription occurs solely within the nucleus, and translation takes place solely in the cytoplasm. Consequently, gene expression regulation in eukaryotes can occur at various stages throughout this process.
 Prokaryotic transcription and translation occur simultaneously in the cytoplasm, and regulation occurs at the level of transcription. In eukaryotes, transcription and translation are physically separated, and gene expression is regulated at many different levels.
Prokaryotic transcription and translation occur simultaneously in the cytoplasm, and regulation occurs at the level of transcription. In eukaryotes, transcription and translation are physically separated, and gene expression is regulated at many different levels.
Differences in prokaryotic and eukaryotic gene regulation

Prokaryotic Gene Regulation
- Prokaryotic organisms have their DNA arranged in a circular chromosome located in the cell's cytoplasm. Genes that are involved in the same biological processes are often grouped together in structures called operons. These operons contain multiple genes that are transcribed into a single mRNA molecule, allowing for coordinated regulation of their expression.
- Prokaryotic cells control gene expression through the actions of activators, which enhance transcription, repressors, which inhibit transcription, and inducers, which deactivate repressors. Below, we'll explore two examples of how these molecules regulate different operons.
The trp Operon: A Repressor Operon
In bacteria like E. coli, the synthesis of amino acids, such as tryptophan, is essential for survival. The genes responsible for tryptophan synthesis are organized in the tryptophan (trp) operon. When tryptophan is available in the environment, E. coli doesn't need to synthesize it, and the trp operon remains inactive. Conversely, when tryptophan is scarce, the trp operon is activated to enable tryptophan synthesis. An operator sequence lies between the promoter and the first trp gene and contains the binding site for the repressor protein. The repressor's conformational change, induced by tryptophan binding, enables it to attach to the trp operator. This binding physically prevents RNA polymerase from binding and transcribing the downstream genes. Therefore, when the cell has sufficient tryptophan, it inhibits further synthesis. The five genes that are needed to synthesize tryptophan in E. coli are located next to each other in the trp operon.
The five genes that are needed to synthesize tryptophan in E. coli are located next to each other in the trp operon.
In the absence of tryptophan, the repressor remains unbound as there's no tryptophan to trigger its activation. Consequently, RNA polymerase can transcribe the operon genes, facilitating tryptophan synthesis when the cell is deficient in this amino acid.
The lac Operon: An Inducer Operon
- The lac operon in E. coli, which is responsible for lactose digestion, is more intricately regulated, involving both a repressor and an activator. While E. coli primarily uses glucose as a food source, it can utilize other sugars like lactose when glucose is scarce. The lac operon encodes the proteins required to break down lactose.
- When lactose is absent, a repressor binds to the operator, preventing RNA polymerase from transcribing the operon. In the presence of lactose, lactose molecules bind to the repressor, removing it from the operator. This action allows RNA polymerase to transcribe the genes necessary for lactose digestion.
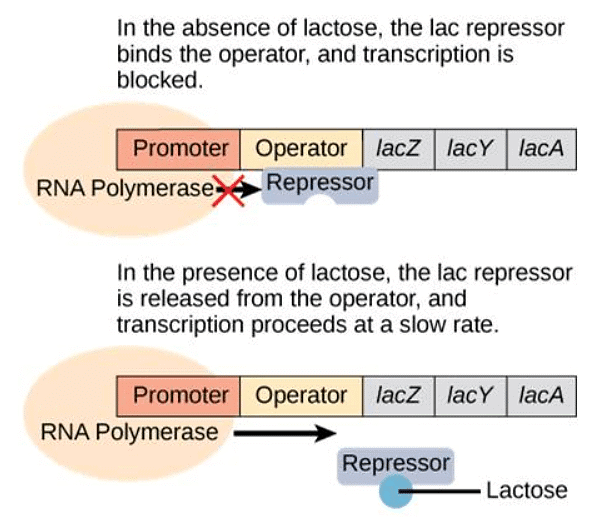 Transcription of the lac operon only occurs when lactose is present. Lactose binds to the repressor and removes it from the operator.
Transcription of the lac operon only occurs when lactose is present. Lactose binds to the repressor and removes it from the operator.
- However, the story becomes more nuanced when only lactose is present. In such cases, the bacterium needs to increase production of the lactose-digesting proteins. This is achieved through an activator protein called catabolite activator protein (CAP). When glucose levels drop, cyclic AMP (cAMP) accumulates in the cell. cAMP then binds to CAP, and the complex attaches to the lac operon promoter. This enhances the binding affinity of RNA polymerase to the promoter, leading to increased transcription of the genes.
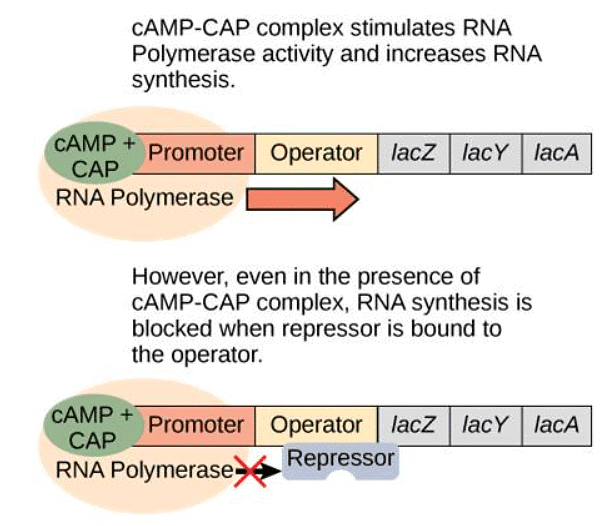 When there is no glucose, the CAP activator increases transcription of the lac operon. However, if no lactose is present, the operon is not activated.
When there is no glucose, the CAP activator increases transcription of the lac operon. However, if no lactose is present, the operon is not activated.
- In summary, for full activation of the lac operon, two conditions must be met: low or absent glucose levels and the presence of lactose. Under these circumstances, the lac operon is transcribed at maximum capacity, conserving energy by only producing the necessary proteins for lactose digestion.
Summary of signals that induce or repress transcription of the lac operon.
Eukaryotic Gene Regulation
In eukaryotes, gene expression is a highly intricate process with various levels of control, differing from the more straightforward regulation in prokaryotes. Unlike prokaryotes, eukaryotic genes are not organized into operons, and each gene is independently regulated. Additionally, eukaryotes possess a significantly larger number of genes compared to prokaryotes. Gene expression in eukaryotes can be regulated at multiple stages, including DNA transcription, mRNA processing, mRNA transport from the nucleus to the cytoplasm, and mRNA binding to ribosomes. To simplify this complexity, gene regulation is often categorized into five levels: epigenetic, transcriptional, post-transcriptional, translational, and post-translational.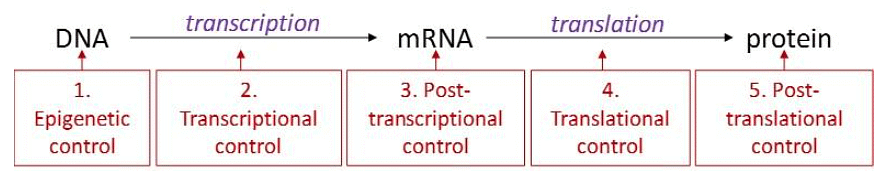 Regulation of gene expression in eukaryotes can occur at five different levels. Here, the Central Dogma is diagrammed with arrows showing where each type of eukaryotic regulation of gene expression interrupts it.
Regulation of gene expression in eukaryotes can occur at five different levels. Here, the Central Dogma is diagrammed with arrows showing where each type of eukaryotic regulation of gene expression interrupts it.
Epigenetic Control of Gene Expression
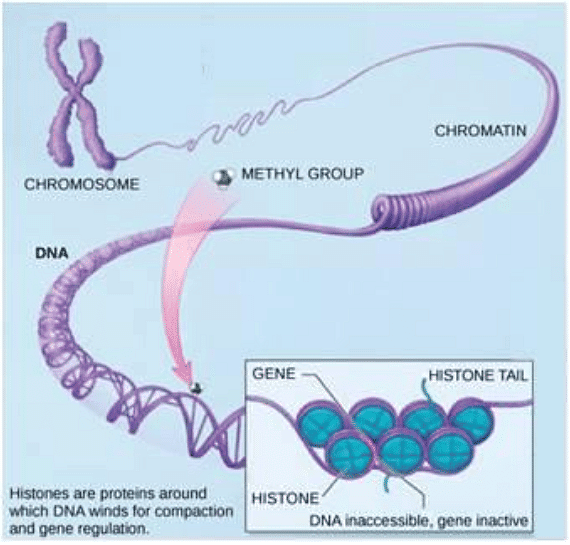
- Epigenetic regulation, the first level of gene expression control, encompasses modifications that don't alter DNA nucleotide sequences and are not permanent. Instead, these modifications modify chromosomal structure, allowing genes to be turned on or off. This level of control involves heritable chemical modifications of DNA and chromosomal proteins.
- One example of epigenetic regulation is DNA methylation, where methyl groups are added to DNA, generally repressing transcription. These methylation patterns can be passed on as cells divide, allowing parental gene expression tendencies to be inherited by offspring. Other heritable chemical modifications of DNA may exist.
- Another example of epigenetic control is the modification of histone proteins, which package and organize DNA into structural units called nucleosome complexes. These histones can move along DNA, altering the chromosomal structure, and enabling or preventing transcription by controlling the access of transcription factors and RNA polymerase to DNA. Chemical tags, like phosphate, methyl, or acetyl groups, attached to histones can open or close chromosomal regions.
Modification of Histone Proteins is an Example of Epigenetic Control
- One prominent illustration of epigenetic regulation is the modification of histone proteins. Histones serve as chromosomal proteins that facilitate the compact winding of DNA, enabling it to fit within the microscopic nucleus of a cell. Given that the human genome contains over three billion nucleotide pairs, and an average chromosome comprises 130 million nucleotide pairs, it's crucial to organize and condense this DNA. This organization ensures that specific segments can be accessed as needed by different cell types.
 In each chromosome, DNA is wound around histone proteins to pack it into the nucleus of a cell. (Credit: modification of work by NIH.)
In each chromosome, DNA is wound around histone proteins to pack it into the nucleus of a cell. (Credit: modification of work by NIH.)
- At the fundamental level of organization, DNA strands wrap around histone proteins. Histones play a critical role in packaging and structuring DNA into units called nucleosome complexes, which can regulate access to DNA regions. When visualized under an electron microscope, this winding of DNA around histone proteins resembles tiny beads on a string. These beads, representing histone proteins, have the ability to move along the string (DNA) and influence the molecule's structure.
 DNA is wrapped around histones to create nucleosomes (a), which control the access of proteins to DNA. When viewed through an electron microscope (b), the nucleosomes look like beads on a string. (Credit “micrograph”: modification of work by Chris Woodcock.)
DNA is wrapped around histones to create nucleosomes (a), which control the access of proteins to DNA. When viewed through an electron microscope (b), the nucleosomes look like beads on a string. (Credit “micrograph”: modification of work by Chris Woodcock.)
- If a particular gene needs to be transcribed, the nucleosomes surrounding the corresponding DNA region can shift along the DNA strand, effectively opening that specific chromosomal region. This opening allows RNA polymerase and other proteins, known as transcription factors, to bind to the promoter region, initiating transcription. Conversely, when a gene should remain inactive or silenced, the histone proteins and DNA undergo different modifications that indicate a closed chromosomal configuration. In this closed state, RNA polymerase and transcription factors cannot access the DNA, preventing transcription.
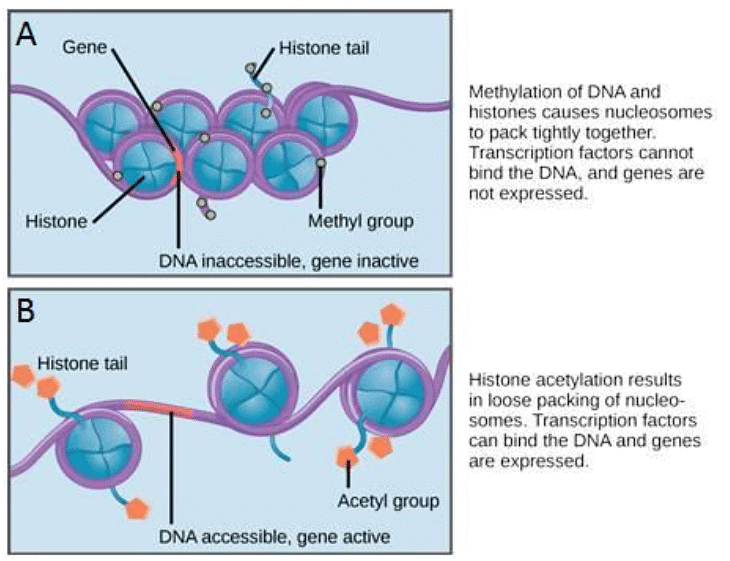 Nucleosomes can slide along DNA. (A) When nucleosomes are spaced closely together, transcription factors cannot bind and gene expression is turned off. (B) When nucleosomes are spaced far apart, transcription factors can bind, allowing gene expression to occur.
Nucleosomes can slide along DNA. (A) When nucleosomes are spaced closely together, transcription factors cannot bind and gene expression is turned off. (B) When nucleosomes are spaced far apart, transcription factors can bind, allowing gene expression to occur.
- The movement of histone proteins is influenced by signals found on these proteins, which serve as "tags." These tags, which take the form of phosphate, methyl, or acetyl groups, can either open or close chromosomal regions. Importantly, these tags are not permanent but can be added or removed as needed. Since DNA carries a negative charge, alterations in histone charge can affect how tightly DNA is wound around the histone proteins. In its unmodified state, histone proteins possess a large positive charge. However, the addition of chemical modifications like acetyl groups reduces this positive charge, altering the level of DNA winding.
Transcriptional Control of Gene Expression
- Transcriptional regulation governs whether mRNA is transcribed from a gene in a specific cell. Transcription in eukaryotes involves RNA polymerase, which requires transcription factors to initiate transcription. Transcription factors are proteins that bind to the promoter and other regulatory sequences to control gene transcription. Unlike prokaryotes, where RNA polymerase alone initiates transcription, in eukaryotes, transcription factors must first bind to the promoter to recruit RNA polymerase.
- Eukaryotic genes have promoter regions immediately upstream of coding sequences. These promoters vary in length and serve as binding sites for transcription factors, facilitating transcription initiation. Within the promoter region, the TATA box, composed of thymine and adenine dinucleotides, is essential. Transcription factors bind to the TATA box, assembling an initiation complex and allowing RNA polymerase to initiate transcription.
- Enhancers, regulatory regions that can be distant from genes, further enhance transcription. Activators bind to enhancers, and when enhancers come into proximity with the promoter, they interact with the transcription initiation complex to enhance transcription. Repressors can also bind to promoter or enhancer regions, blocking transcription. Both activators and repressors respond to external cues to determine gene expression.
The Promoter and Transcription Factors
- In eukaryotic genes, the promoter region is situated just upstream of the coding sequence. This region's length can vary widely, ranging from a few nucleotides to hundreds, and is specific to each gene. The extent of the promoter's length directly impacts the available space for proteins to bind, resulting in substantial variations in gene expression control among different genes. The primary function of the promoter is to serve as a binding site for transcription factors, which regulate the initiation of transcription (as depicted in Figure 17.10, top).
- Located within the promoter region, immediately upstream of the transcriptional start site, is the TATA box. The TATA box consists of a repeated sequence of thymine and adenine dinucleotides (abbreviated as TATA repeats). Transcription factors attach themselves to the TATA box, facilitating the assembly of an initiation complex. Once this complex is formed, RNA polymerase binds to the upstream sequence and undergoes phosphorylation. This phosphorylation event causes a portion of the protein to detach from the DNA, activating the transcription initiation complex. Consequently, RNA polymerase is correctly positioned to commence transcription (illustrated in Figure 17.10, top).
 Each gene has a promoter upstream of the coding sequence. The promoter binds to transcription factors and helps RNA polymerase to bind and start transcription. Bottom. Many genes also have upstream enhancers. Enhancers bind activators, bend around, and help RNA polymerase start transcription.
Each gene has a promoter upstream of the coding sequence. The promoter binds to transcription factors and helps RNA polymerase to bind and start transcription. Bottom. Many genes also have upstream enhancers. Enhancers bind activators, bend around, and help RNA polymerase start transcription.
Enhancers and Repressors
- In certain eukaryotic genes, specific regions contribute to enhancing transcription. These regions, known as enhancers, are not necessarily located in close proximity to the genes themselves; they can be positioned thousands of nucleotides away from the gene locus. Enhancers can be found upstream, within the coding region, or downstream of a gene. These enhancers serve as binding sites for activator proteins. When an enhancer is located at a considerable distance from a gene, the DNA undergoes structural changes, causing the enhancer to come into proximity with the promoter region. This allows for interactions between activators and the transcription initiation complex, effectively promoting transcription (as illustrated in Figure 17.10, bottom).
- Similar to prokaryotic cells, eukaryotic cells possess mechanisms to inhibit transcription. Transcriptional repressor proteins can attach themselves to either promoter or enhancer regions, effectively obstructing the process of transcription. Both activator and repressor proteins respond to external stimuli, determining which genes should be expressed in response to various cues and conditions.
Post-transcriptional Control of Gene Expression
Post-transcriptional regulation occurs after mRNA transcription but before translation. It involves mRNA processing, transport, and binding to ribosomes. Alternative RNA splicing allows different combinations of introns and sometimes exons to be removed, producing various protein products from one gene. RNA-binding proteins (RBPs) can bind to mRNA regions, altering mRNA stability. miRNAs, short RNA molecules, can also bind to mRNA, leading to rapid mRNA degradation.
Alternative RNA splicing
- It is important to note that eukaryotic cell RNA primary transcripts often contain segments known as introns, which are typically removed before the process of translation can take place.
- Alternative RNA splicing represents a mechanism that permits the selective removal of different combinations of introns, and sometimes exons, from the primary transcript (depicted in Figure 17.11). This process results in the generation of various protein products derived from a single gene.
- Alternative splicing serves as a means of regulating gene expression, allowing the production of different protein variants in distinct cells or at varying times within the same cell. It has been established that alternative splicing is a prevalent mechanism of gene regulation in eukaryotic organisms, with as many as 70 percent of human genes being expressed as multiple protein isoforms through this process.
 Before a RNA can be translated, introns must be removed by splicing. Pre-mRNA can be alternatively spliced to create different proteins.
Before a RNA can be translated, introns must be removed by splicing. Pre-mRNA can be alternatively spliced to create different proteins.
Control of RNA Stability
- An additional form of post-transcriptional control pertains to the stability of mRNA within the cytoplasm. The longer mRNA persists in the cytoplasm, the greater the opportunity for translation to occur, leading to increased protein production. Several factors influence mRNA stability, with one significant factor being the length of its poly-A tail.
 The protein-coding region of mRNA is flanked by 5′ and 3′ untranslated regions (UTRs). RNA-binding proteins at the 5′ or 3′ UTR influence the stability of the RNA molecule.
The protein-coding region of mRNA is flanked by 5′ and 3′ untranslated regions (UTRs). RNA-binding proteins at the 5′ or 3′ UTR influence the stability of the RNA molecule.
- RNA-binding proteins (RBPs), specialized proteins, have the capability to attach to regions of RNA situated immediately preceding or following the protein-coding region. These sections in the RNA, which do not undergo translation into protein, are referred to as untranslated regions (UTRs). Specifically, the portion just preceding the protein-coding region is known as the 5' UTR, while the segment following the coding region is called the 3' UTR (illustrated in Figure 17.12). The interaction between RBPs and these regions can either enhance or diminish the stability of the RNA molecule, contingent on the specific RBP involved.
- Another regulatory element involves microRNAs (miRNAs), which are brief RNA molecules, typically consisting of 21–24 nucleotides. MiRNAs are initially synthesized in the nucleus as longer pre-miRNAs and subsequently processed into mature miRNAs by an enzyme called dicer. MiRNAs, in conjunction with a ribonucleoprotein complex known as the RNA-induced silencing complex (RISC), participate in the swift degradation of the target mRNA.
Translational Control of Gene Expression
Translational regulation controls protein synthesis after mRNA has been transported to the cytoplasm. mRNA stability significantly influences translation. Additionally, translation initiation can be regulated by mRNA-ribosome binding. For proteins destined for the endoplasmic reticulum (ER), a signal sequence on the mRNA leads to translational pausing until the mRNA-ribosome complex reaches the ER.
Post-translational Control of Gene Expression
- The ultimate tier of gene expression control in eukaryotes is post-translational regulation, which involves modifying a protein once it has been synthesized, thereby influencing its activity. A notable example of post-translational regulation pertains to enzyme inhibition. When an enzyme is no longer required, it can be inhibited by either a competitive or allosteric inhibitor, which prevents its binding to a substrate. Importantly, this inhibition is reversible, allowing for the reactivation of the enzyme at a later time. This process proves more efficient than degrading the enzyme when it is no longer needed, followed by synthesizing it anew when required.
- Furthermore, the function and/or stability of proteins can be regulated through the addition of functional groups like methyl, phosphate, or acetyl groups. In some cases, these modifications dictate the cellular localization of a protein, determining whether it is found in the nucleus, cytoplasm, or attached to the plasma membrane.
- The attachment of a ubiquitin group to a protein serves as a marker indicating that the protein's lifecycle has concluded. Ubiquitin effectively functions as a flag, signifying the protein's readiness for degradation. Tagged proteins are transported to a cellular organelle known as the proteasome, where they undergo degradation (as depicted in Figure 17.13). This means that one mechanism for regulating gene expression involves manipulating the duration of a protein's presence within the cell.
 Proteins with ubiquitin tags are marked for degradation within the proteasome.
Proteins with ubiquitin tags are marked for degradation within the proteasome.
|
179 videos|140 docs
|
FAQs on Regulation of Gene Expression - Botany Optional for UPSC
| 1. What is gene expression regulation? |  |
| 2. How is gene expression regulated in prokaryotes? |  |
| 3. What are some mechanisms of gene expression regulation in eukaryotes? |  |
| 4. How can gene expression be influenced by environmental factors? |  |
| 5. Why is the regulation of gene expression important? |  |

|
Explore Courses for UPSC exam
|

|

















