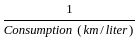CSIR NET Life Sciences Mock Test - 2 - UGC NET MCQ
30 Questions MCQ Test - CSIR NET Life Sciences Mock Test - 2
Percentage of six different type of garments in two stores are given below:
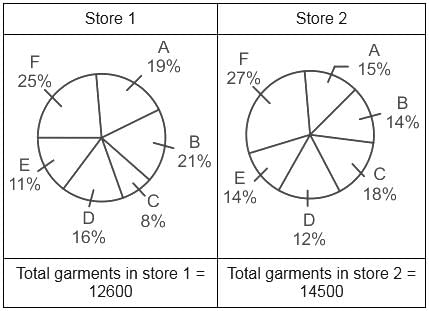
Which type of garments has the minimum difference between the two stores?

Which type of garments has the minimum difference between the two stores?
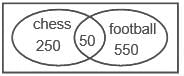
In a school of 1000 students, 300 students play chess and 600 students play football. If 50 students play both chess and football, the number of students who play neither is
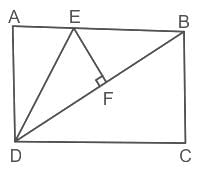
In the given figure, ABCD is a rectangle with AD = 4 units and AE = EB. EF is perpendicular to DB and is half of DF. If the area of the triangle DEF is 5 sq. units, then what is the area of ABCD?

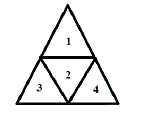
In the figure shown above, what is the maximum number of different ways in which 8 identical balls can be placed in the small triangles 1,2,3 and 4 such that each triangle contains at least one ball ?
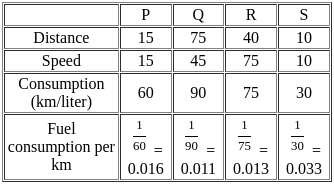
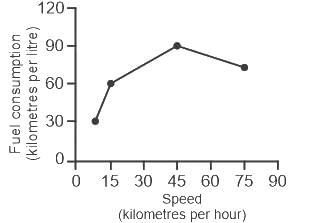
The fuel consumed by a motorcycle during a journey while traveling at various speeds a indicated in the graph below.
The distances covered during four laps of the journey are listed in the table below:
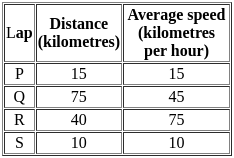
From the given data, we can conclude that the fuel consumed per kilometre was least during the lap
Among five girls standing side by side, Leela has exactly one girl to her left, Alice is just right of Prerna, and there are at least two girls between Radha and Zarina. The girl in the middle is
A sample of DNA is found to have the base composition (mole ratio) of A = 40, T = 22, G = 21 and C = 17. This suggests:
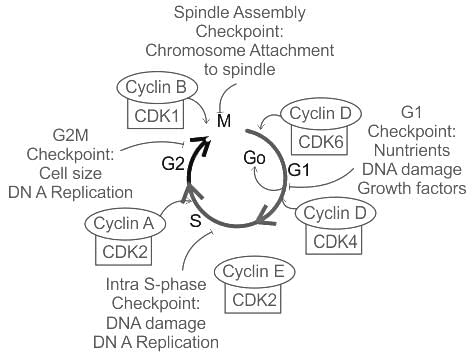
During the cell cycle, the G2/M checkpoint ensures that cells do not enter mitosis before DNA replication is complete and that any DNA damage is repaired. Which of the following protein complexes is crucial for the regulation of this checkpoint?
Which one of the following method can be used to treat crude sewage and nitrify secondary effluent?
Which of the following techniques can be used to generate transgenic Drosophila strains?
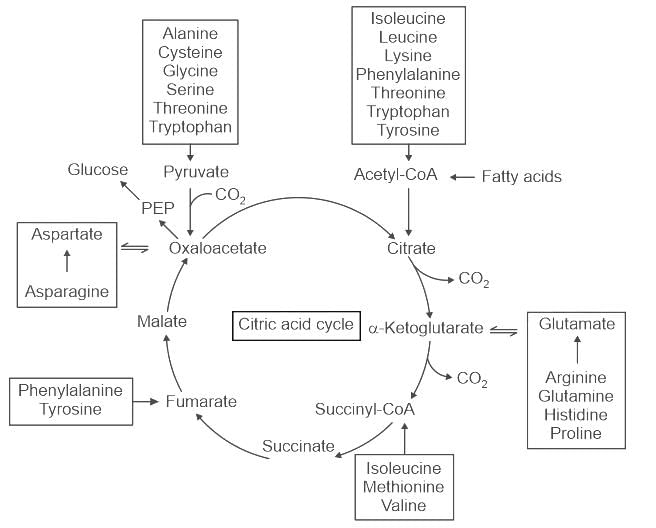
The common metabolic precursor of amino acids such as Proline and Glutamine is
Which one of the following four statements is true regarding nitrogen fixation by bacteria?
The protein concentration and enzyme activity in 100 mL of a cell free extract is 5 mg/mL and 2 units/mL, respectively. After multiple steps of purification, the final 10 mL fraction contains 4 mg/mL of protein and 15 units/mL of enzyme activity. The fold purification and percentage recovery, respectively is:
Brassica juncea has bisexual flowers.
A mutation in the mitochondria leads to cytoplasmic male sterility (CMS). CMS can be restored by a restorer of fertility gene ( Rf ) which is a nuclear gene.
Fertility restoration is a dominant phenotype.
A CMS line is crossed to a homozygous Rf line. The obtained F1 progeny is selfpollinated. What percentage of F2 progeny will be male sterile?
Which of the following statements about gene therapy are CORRECT?
[P] Affected individuals, but not their progeny, can be cured through germline gene therapy
[Q] Affected individuals, as well as their progeny, can be cured through germline gene therapy
[R] Affected individuals, but not their progeny, can be cured through somatic gene therapy
[S] Affected individuals, as well as their progeny, can be cured through somatic gene therapy
Consider the following statements related to protein sorting and targeting to cell organelles:
1. Signal recognition particle (SRP) is essential for the targeting of specific proteins to the endoplasmic reticulum (ER).
2. Proteins destined for the nucleus are transported through nuclear pore complexes, and their sorting relies on a short amino acid sequence known as a nuclear localization signal (NLS).
3. Mitochondrial proteins are synthesized with a mitochondrial targeting sequence that is recognized by receptors on the mitochondrial surface.
4. Proteins bound for the peroxisome typically contain a peroxisomal targeting signal (PTS1), which is recognized by the cytosolic receptor Pex5p.
Based on the above statements, which of the following options is correct?
Regional heterothermy is seen in the following insects
i. Honey bees
ii. Termites
iii. Bumble bees
iv. Moths
The correct combination is
In a genetic cross between plants bearing violet flowers and green seeds (VVGG), and white flower and yellow seeds (vvgg), the following phenotypic distribution was obtained in the F2 progeny (assume both parents to be pure breeding for both the traits, and self-cross at F1 generation):
i) 2340 plants with violet flowers and green seeds
ii) 47 plants with violet flowers and yellow seeds
iii) 43 plants with white flowers and green seeds
iv) 770 plants with white flowers and yellow seeds
Which one of the following interpretations explains the above phenotypic distribution?
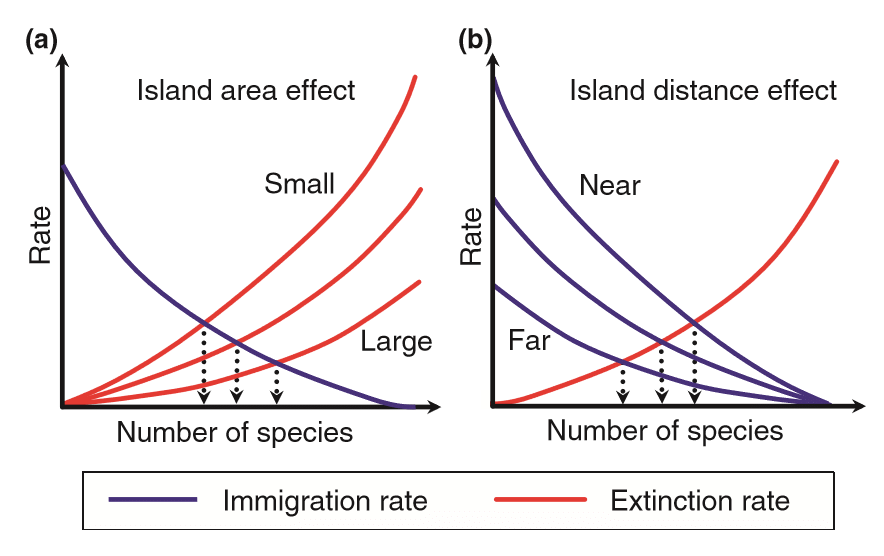
Biogeography is the study of the geographical distribution of species and the reason for their pattern of distribution. Ecologists Robert MacArthur and E.O. Wilson, proposed the equilibrium theory of Island biogeography. By 'islands' we mean not only oceanic islands, but also habitat islands on land, such as the peaks of mountains and isolated springs in the desert. Certain statement were made regarding Island biogeography.
A. The theory proposed that the number of species on any island is determine by balance between the rate at which new species colonize it and the rate at which population of established species become extinct.
B. The theory proposed that the number of species on any island is determine by balance between the rate at which old species colonize it and the rate at which population of established species become diversify.
C. Islands that are more isolated are less likely to receive immigrants than islands that are less isolated.
D. Small islands have slow extinction rates.
Select the correct combination of statement.
Choose the correct statements.
P. Tc cells recognize antigen with class I MHC molecules on target cells.
Q. Endogenous antigens are degraded into peptides within the cytosol by proteasomes.
R. Exogenous antigens are internalized and degraded within the acidic endocytic compartments.
S. Presentation of nonpeptide antigens derived from bacteria involves the class I-like CD1 molecules.



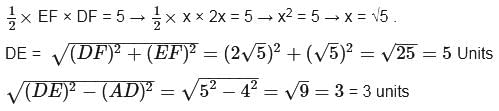
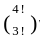 = 4 w a y s ) ;
= 4 w a y s ) ;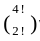 = 12 w a y s ) ;
= 12 w a y s ) ;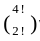 × 2 ! = 6 w a y s ) ;
× 2 ! = 6 w a y s ) ;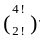 = 12 w a y s ) and
= 12 w a y s ) and