CSIR NET Life Sciences Mock Test - 3 - UGC NET MCQ
30 Questions MCQ Test - CSIR NET Life Sciences Mock Test - 3
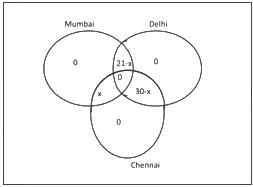
Suppose three meetings of a group of professors were arranged in Mumbai, Delhi and Chennai. Each professor of the group attended exactly two meetings. 21 professors attended Mumbai meeting, 27 attended Delhi meeting and 30 attended Chennai meeting. How many of them attended both the Chennai and Delhi meetings?
The sum of any three consecutive natural numbers where first number is even number is always divisible by?
There are two buckets A and B. Initially A has 2 litres of water and B is empty. At every hour 1 litre of water is transferred from A to B followed by returning 1/2 litre back to A from B half an hour later. The earliest A will get empty is in:
Two identical cube shaped dice each with faces numbered 1 to 6 are rolled simultaneously. The probability that an even number is rolled out on each dice is:
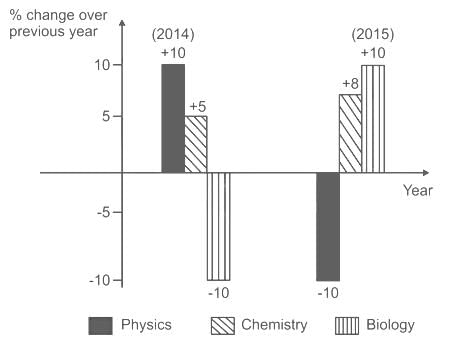
Which of the following inferences can be drawn from the above graph?
For any four consecutive decimal digits, the largest value of the product of the sum of any two and the sum of the other two is
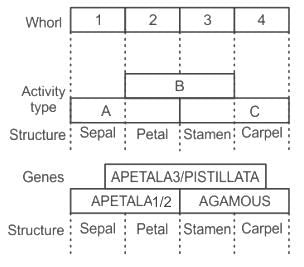
In agamous (ag) mutants, which of the floral whorls is affected:
Which protein family is primarily involved in regulating apoptosis during the formation of digits in vertebrate limb development?
In the process of salting out, why do added ions preferentially interact with water molecules instead of the protein?
Assertion [a]: The cardiovascular organization called double circulation provides vigorous flow of blood to the brain, muscles, and other organs.
Reason [r]: The blood is pumped a second time after it loses pressure in the capillary beds of the lungs or skin.
A writer named "x" has written the first description of a plant variety. Later, author "y" moved the species to a different genus. The new combination's source citation will then be:
Choose the statement that is not correct for the cytoskeleton protein actin
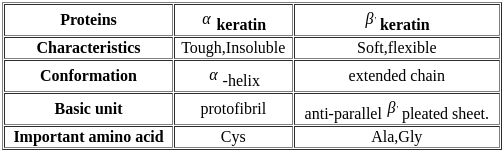
α-Keratin’s chemical composition is notably rich in which types of amino acids?
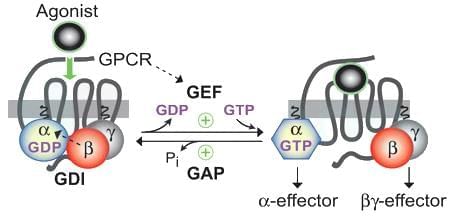
Which of the following processes turn - on and turn - off the alpha subunit of a heterotrimeric G - protein?
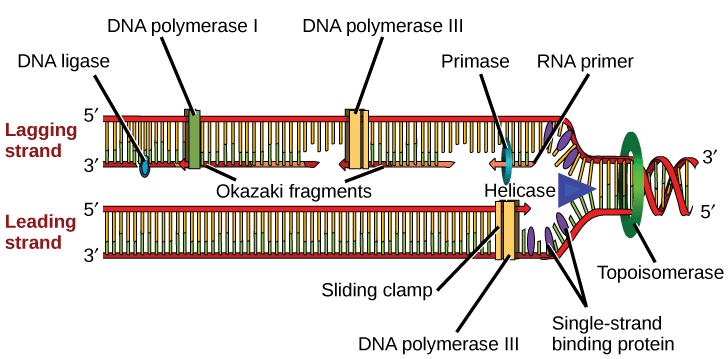
During the initiation of DNA replication in prokaryotes, which of the following is the correct sequence of events?
Which of the following vitamins becomes part of a high-energy metabolite in the cell?
A DNA molecule is completely transcribed into messenger RNA by an RNA polymerase. The base composition of the DNA template strand is G = 24.1%; C = 18.5%; A = 24.6%; T = 32.8%. The base composition of the newly synthesized RNA molecule is:
Consider the following statements regarding the cell cycle and cell division:
- The G1 checkpoint, also known as the restriction point, assesses cell size, nutrients, and DNA integrity before proceeding with DNA replication.
- During the S phase, DNA is replicated, and the replication machinery ensures that each daughter cell will receive an exact copy of the genome.
- The G2/M checkpoint ensures that DNA replication is completed successfully and checks for DNA damage. If the cell passes this checkpoint, it is committed to mitosis.
- The spindle assembly checkpoint (SAC) occurs during the G2 phase and ensures that all chromosomes are properly aligned on the spindle before anaphase onset.
Which of the following is the most accurate correlation of these statements to the cell cycle phases and their checkpoints?
The table given below provides a list of seed or fruit characteristics and plant genera:
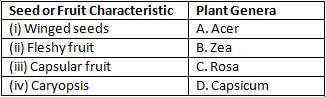
Which one of the following options correctly matches the plant genera to seed or fruit characteristics?
Based on the statement, which of the following statements is correct?
India has currently 17 biosphere reserves representing different ecosystems. These conservation areas significantly differ from the conventional protected areas of the country. Identify the correct combination of attributes (A to G) that best explains the concept of biosphere reserve.
(A) Conservation,
(B) Education,
(C) Human habitation allowed,
(D) Human habitation not allowed,
(E) Strong legal back-up,
(F) No supporting act,
(G) Research.
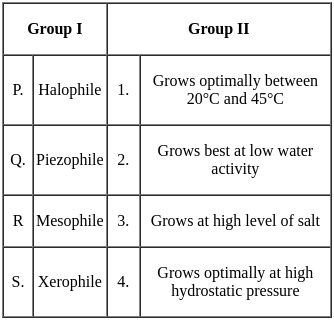
Match the type of bacteria in Group I with their respective growth properties in Group II.
Statement 1: In phylogenetic analysis of scorpion species Mesobuthus tamulus, single nucleotide polymorphisms (SNPs) were used to infer ancestral relationships.
Statement 2: The principle of parsimony suggests that the ancestral nucleotide at a given node will minimize the number of changes required to explain observed SNP differences in descendants.
Statement 3: If three individuals show A, A, and T at a specific locus, the most parsimonious ancestral nucleotide at the corresponding internal node is likely to be A.
Which of the following statements are correct?
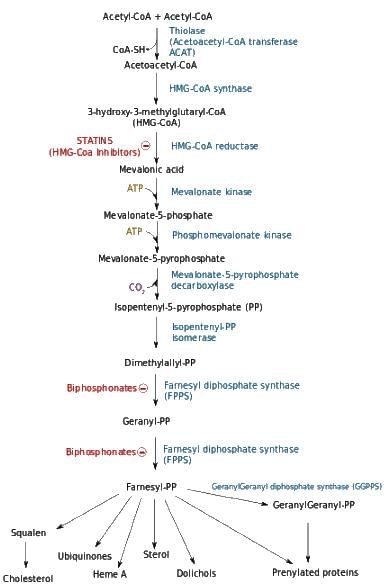
Arrange the following in proper sequence in the biosynthetic pathway.
I. Geranyl pyrophosphate is converted to farnesyl pyrophosphate
Il. Geranylgeranyl pyrophosphate is converted to copalyl pyrophosphate
III. Mevalonic acid is converted to mevalonic acid pyrophosphate
IV. Dimethylallyl pyrophosphate is converted to geranyl pyrophosphate
V. Isopentenyl pyrophosphate is converted to dimethylallyl pyrophosphate
The following statements are made about the killing of tumor cells by natural killer (NK) cells:
A. Tumor cells often downregulate MHC-I molecules.
B. NK cells recognize the absence of MHC-I molecules on target cells.
C. NK cells recognize tumor antigens presented on MHC-II molecules.
D. NK cells kill tumor cells through the release of perforins and granzymes.
Which one of the following options represents the combination of all correct statements?
A bacterial cell elongates at a rate of 1.2 µm per minute under optimal conditions. Assuming that the cell wall is made entirely of peptidoglycan, how many N-acetylglucosamine (NAG) residues must be added per second to accommodate this rate of growth? The length of one NAG residue in peptidoglycan is about 0.5 nm.
Following are certain facts about the effect of abscisic acid (ABA) on the development and physiological effect of plants:
A. ABA promotes leaf senescence independent of ethylene
B. ABA promotes shoot growth and inhibits root growth at low water potential
C. ABA inhibits gibberellin induced enzyme production
D. Seed dormancy is controlled by ratio of ABA and gibberellin
Which of the following combinations based on above statements is correct?
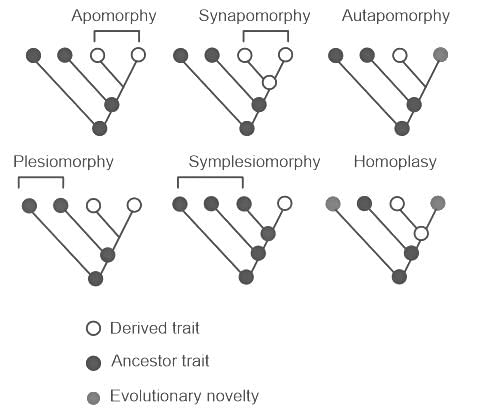
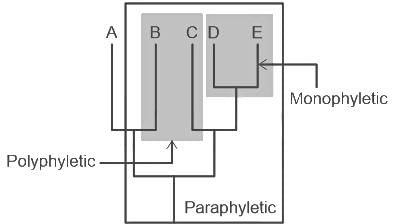
Three living species X, Y and Z share a common ancestor T, as do extinct species U and V. A grouping that consists of species T, X, Y, and Z (but not U or V) makes up:



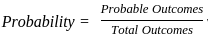
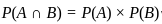
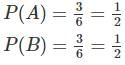
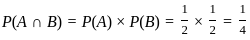
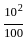 = 1% loss
= 1% loss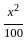 %
%