All Exams >
NEET >
Biology Class 12 >
All Questions
All questions of Molecular Basis of Inheritance for NEET Exam
Match column with column II and select the correct option from the given codes.

- a)A-(i), B-(i), C-(iii)
- b)A-(i), B-(iii), C-(ii)
- c)A-(iii), B-(i), C-(ii)
- d)A-(ii), B-(iii), C-(i)
Correct answer is option 'B'. Can you explain this answer?
Match column with column II and select the correct option from the given codes.


a)
A-(i), B-(i), C-(iii)
b)
A-(i), B-(iii), C-(ii)
c)
A-(iii), B-(i), C-(ii)
d)
A-(ii), B-(iii), C-(i)

|
Lead Academy answered |
DNA Replication: DNA replication is the process by which a cell makes an identical copy of its DNA. It occurs during the cell cycle to ensure that each daughter cell receives a complete set of genetic instructions. DNA replication is a highly accurate process in which the DNA double helix is unwound, and each strand serves as a template for the synthesis of a complementary strand.
Transcription: Transcription is the process by which a segment of DNA is used as a template to synthesize a complementary RNA molecule. It is the first step in the central dogma of molecular biology, which describes how genetic information is used to produce proteins. In transcription, an enzyme called RNA polymerase reads the DNA template and assembles a single-stranded RNA molecule, known as messenger RNA (mRNA), by matching RNA nucleotides to the DNA template.
Translation: Translation is the process in which the information contained in an mRNA molecule is used to synthesize a protein. This process occurs at ribosomes, cellular structures that serve as the protein-manufacturing factories. During translation, the mRNA is read in sets of three nucleotides called codons. Each codon corresponds to a specific amino acid, the building blocks of proteins.
The structure in chromatin seen as 'beads-on' string' when viewed under electron microscope are called- a)nucleotides
- b)nuclesides
- c)histone octamer
- d)nucleosomes
Correct answer is option 'D'. Can you explain this answer?
The structure in chromatin seen as 'beads-on' string' when viewed under electron microscope are called
a)
nucleotides
b)
nuclesides
c)
histone octamer
d)
nucleosomes
|
|
Meera Singh answered |
Nucleosomes constitute the repeating unit of a structure in nucleus called chromatin, thread-like stained (coloured) bodies seen in nucleus. The nucleosomes in chromatin are seen as 'beads-on-string' structure when viewed under electron microscope.
Select the correct match of enzyme with its related function- a)DNA polymerase - Synthesis of DNA Strands
- b)Helicase - Unwinding of DNA helix
- c)Ligase - Joins together short DNA segments
- d)All of these
Correct answer is option 'D'. Can you explain this answer?
Select the correct match of enzyme with its related function
a)
DNA polymerase - Synthesis of DNA Strands
b)
Helicase - Unwinding of DNA helix
c)
Ligase - Joins together short DNA segments
d)
All of these

|
Stepway Academy answered |
DNA Polymerase: Synthesis of DNA Strands DNA polymerase is an enzyme responsible for the synthesis of new DNA strands during DNA replication. It adds complementary nucleotides to the existing DNA template strand, following the base-pairing rules (A with T and C with G), to create two identical DNA molecules from one original strand. DNA polymerase is essential for the accurate and efficient replication of DNA and plays a crucial role in maintaining the genetic integrity of an organism.
Helicase: Unwinding of DNA Helix Helicase is an enzyme that unwinds the double-stranded DNA helix by breaking the hydrogen bonds between the complementary base pairs. This unwinding action is a crucial step in DNA replication, as it separates the two DNA strands and exposes the template strands for replication by DNA polymerase. Helicase ensures that the DNA molecule can be duplicated and transcribed for various cellular processes like replication, transcription, and repair.
Ligase: Joins Together Short DNA Segments DNA ligase is an enzyme that plays a key role in the process of DNA replication and repair. It is responsible for joining together the short DNA fragments, known as Okazaki fragments, on the lagging strand during DNA replication. These fragments are synthesized in the opposite direction of the replication fork. DNA ligase catalyzes the formation of phosphodiester bonds between the sugar-phosphate backbones of adjacent DNA fragments, creating a continuous and functional DNA strand. In addition to replication, DNA ligase is also involved in DNA repair processes, ensuring the integrity of the genetic material.
Choose the correct answer from the alternatives given:
What would be the base sequence of RNA transcript obtained from the given DNA segment?
5-G C A T T C G G C T A G T A A C-3 Coding strand of DNA
3-C G T A A G C C G A T C A T T G-5 Non-coding strand of DNA- a)5′−GCAUUCGGCUAGUAAC−3′
- b)5′−CGUAAGCCGAUCAUUG−3′
- c)5′−GCATTCGGCTAGTAAC−3′
- d)3′−CGTAAGCCGATCATTG−5′
Correct answer is option 'A'. Can you explain this answer?
Choose the correct answer from the alternatives given:
What would be the base sequence of RNA transcript obtained from the given DNA segment?
5-G C A T T C G G C T A G T A A C-3 Coding strand of DNA
3-C G T A A G C C G A T C A T T G-5 Non-coding strand of DNA
What would be the base sequence of RNA transcript obtained from the given DNA segment?
5-G C A T T C G G C T A G T A A C-3 Coding strand of DNA
3-C G T A A G C C G A T C A T T G-5 Non-coding strand of DNA
a)
5′−GCAUUCGGCUAGUAAC−3′
b)
5′−CGUAAGCCGAUCAUUG−3′
c)
5′−GCATTCGGCTAGTAAC−3′
d)
3′−CGTAAGCCGATCATTG−5′
|
|
Priya Menon answered |
Since the two strands have opposite polarity and the DNA-dependent RNA polymerase also catalyse the polymerisation in only one direction, that is, 5'→3' the strand that has the sequence same as RNA (except thymine at the place of uracil ) is displaced during transcription. This strand (which does not code for anything ) is referred to as coding strand.
Choose the correct answer from the alternatives given :
If the DNA of a virus is labelled with 32P and the protein of the virus is labelled with 35S, after transduction which molecule(s) would be present inside the bacterial cells?
- a)32P only
- b)35S only
- c)Both (a) and (b)
- d)None of these
Correct answer is option 'A'. Can you explain this answer?
Choose the correct answer from the alternatives given :
If the DNA of a virus is labelled with 32P and the protein of the virus is labelled with 35S, after transduction which molecule(s) would be present inside the bacterial cells?
If the DNA of a virus is labelled with 32P and the protein of the virus is labelled with 35S, after transduction which molecule(s) would be present inside the bacterial cells?
a)
32P only
b)
35S only
c)
Both (a) and (b)
d)
None of these
|
|
Lavanya Menon answered |
- Bacteria which were infected with viruses that had radioactive DNA (incorporated with 32P) were radioactive, indicating that DNA was the material that passed from the virus to the bacteria.
- Bacteria that were infected with viruses that had radioactive proteins (incorporated with 35S) were not radioactive. DNA is therefore, the genetic material that is passesd from virus to bacteria.
Choose the correct answer from the alternatives given:
Watson and Crick (1953) proposed DNA double helix model and won the Nobel Prize; their model of DNA was based on
(i) X-ray diffraction studies of DNA done by Wilkins and Franklin.
(ii) Chargaffs base equivalence rule.
(iii) Griffiths transformation experiment.
(iv) Meselson and Stahls experiment.- a)(i), (ii) and (iv)
- b)(i) and (ii)
- c)(iii) and (iv)
- d)(i), (ii), (iii) and (iv)
Correct answer is option 'B'. Can you explain this answer?
Choose the correct answer from the alternatives given:
Watson and Crick (1953) proposed DNA double helix model and won the Nobel Prize; their model of DNA was based on
(i) X-ray diffraction studies of DNA done by Wilkins and Franklin.
(ii) Chargaffs base equivalence rule.
(iii) Griffiths transformation experiment.
(iv) Meselson and Stahls experiment.
Watson and Crick (1953) proposed DNA double helix model and won the Nobel Prize; their model of DNA was based on
(i) X-ray diffraction studies of DNA done by Wilkins and Franklin.
(ii) Chargaffs base equivalence rule.
(iii) Griffiths transformation experiment.
(iv) Meselson and Stahls experiment.
a)
(i), (ii) and (iv)
b)
(i) and (ii)
c)
(iii) and (iv)
d)
(i), (ii), (iii) and (iv)
|
|
Anjali Sharma answered |
In 1953, James Watson and Francis Crick based on the X-ray diffraction data produced by Maurice Wilkins and Rosalind Franklin, proposed double helix model for the structure of DNA. One of the hallmarks of their proposition was base pairing between two strands of polynucleotide chains. However, this proportion was also based on the observation of Erwin Chargaff (1950) that for a double stranded DNA, the ratios between Adenine and Thymine and Guanine and Cytosine are constant and equals one.
The process of copying genetic information from one strand of DNA to RNA is termed as __________- a)replication
- b)transcription
- c)translation
- d)reverse transcription
Correct answer is option 'B'. Can you explain this answer?
The process of copying genetic information from one strand of DNA to RNA is termed as __________
a)
replication
b)
transcription
c)
translation
d)
reverse transcription
|
|
Ananya Das answered |
The process of copying genetic information from one strand of the DNA into RNA is termed as transcription. Unlike the process of replication, which once sets in, the total DNA of an organism gets duplicated, in transcription only a segment of DNA and only one of the strands is copied into RNA.
Choose the correct answer from the alternatives given:
The year 2003 was celebrated as the 50th anniversary of discovery of- a)Transposons by Barbara Mc Clintock
- b)Structure of DNA by Watson and Crick
- c)Mendels laws of inheritance
- d)Bio technology by Kary Mullis
Correct answer is option 'B'. Can you explain this answer?
Choose the correct answer from the alternatives given:
The year 2003 was celebrated as the 50th anniversary of discovery of
The year 2003 was celebrated as the 50th anniversary of discovery of
a)
Transposons by Barbara Mc Clintock
b)
Structure of DNA by Watson and Crick
c)
Mendels laws of inheritance
d)
Bio technology by Kary Mullis
|
|
Suresh Iyer answered |
Watson and Crick (1953) worked out the first correct double helix model from the X-ray photographs of Wilkins and Franklin. They were awarded Nobel Prize for the same in 1962.
Which of the following phenomena was experimentally proved by Meselson and Stahl?- a)Transformation
- b)Transduction
- c)Semi-conservative DNA replication
- d)Central dogma
Correct answer is option 'C'. Can you explain this answer?
Which of the following phenomena was experimentally proved by Meselson and Stahl?
a)
Transformation
b)
Transduction
c)
Semi-conservative DNA replication
d)
Central dogma
|
|
Anjali Sharma answered |
The Meselson and Stahl experiment was an experiment to prove that DNA replication was semi-subsequently and it was first shown in Escherichia coli and subsequently in higher organisms, such as plants and human cells. Semi-conservative replication means that when the double stranded DNA helix was replicated, each pf the two double stranded DNA helices consisted of one strand coming from the parental helix and one is newly synthesised.
During expression of an operon, RNA polymerase binds to- a)structural gene
- b)regulator gene
- c)operator
- d)promoter
Correct answer is option 'D'. Can you explain this answer?
During expression of an operon, RNA polymerase binds to
a)
structural gene
b)
regulator gene
c)
operator
d)
promoter
|
|
Raghav Bansal answered |
Promoter gene acts as an initiation signal which functions as recognition center for RNA polyrmerase provided the operator gene is switched on. RNA polymerase is bound to the promoter gene. When the operator gene is functional, the polymerase moves over it and reaches the structural genes to perform transcription.
To prove that DNA is the genetic material, which radioactive isotopes were used by Hershey and Chase(1952) in their experiments?- a)35S and 15N
- b)32P and 35S
- c)32P and 15N
- d)14N and 5N
Correct answer is option 'B'. Can you explain this answer?
To prove that DNA is the genetic material, which radioactive isotopes were used by Hershey and Chase(1952) in their experiments?
a)
35S and 15N
b)
32P and 35S
c)
32P and 15N
d)
14N and 5N
|
|
Vivek Patel answered |
Hershey and Chase experiment is based on the fact that DNA contains phosphorus and similarly, sulphur is present in proteins but not in DNA. They incorporated radioactive isotope of phosphorus (32P) into phage DNA and that of sulphur (35S) into proteins of a separate phage culture. Viruses grown in the presence of radioactive phosphorus contained radioactive DNA but not radioactive protein because DNA contains phosphorus but proteins do not, Similarly, viruses grown on radioactive sulphur contain radioactive- protein but not-radioactive DNA because DNA does not contain sulphur.
In a DNA strand, the nucleotides are linked together by- a)glycosidic bonds
- b)phosphodiester bonds
- c)peptide bonds
- d)hydrogen bonds
Correct answer is option 'B'. Can you explain this answer?
In a DNA strand, the nucleotides are linked together by
a)
glycosidic bonds
b)
phosphodiester bonds
c)
peptide bonds
d)
hydrogen bonds
|
|
Vivek Patel answered |
In a DNA strand, the nucleotides are linked together by 3'-5' phosphodiester linkage (bonds ) to form a dinucleotide. More nucleotides can be joined in such a manner to from a polynucleotide chain.
In transcription in eukaryotes, heterogenous nuclear RNA (hn RNA) is transcribed by- a)RNA polymerase I
- b)RNA polymerase II
- c)RNA polymerase III
- d)All of these
Correct answer is option 'B'. Can you explain this answer?
In transcription in eukaryotes, heterogenous nuclear RNA (hn RNA) is transcribed by
a)
RNA polymerase I
b)
RNA polymerase II
c)
RNA polymerase III
d)
All of these
|
|
Meera Singh answered |
Eukaryotes have three RNA polymerases. RNA polymerase I is located in the nucleolus and transcribes for RNAs (28S, 18S and 5.8S) whereas RNA polymerase II is localised in the nucleoplasm and used for hnRNA, mRNA and RNA polymerase Ill is localised in the nucleus, possibly the nucleolar-nucleoplasm interface and transcribes for tRNA, 5S, RNA and nRNAs.
The fully processed hnRNA is called (i) and is transported out of the (ii) into (iii) for translation.- a)(i) mRNA, (ii) Nucleus, (iii) Cytoplasm
- b)(i) mRNA, (ii) Cytoplasm, (iii) Nucleus
- c)(i) tRNA, (ii) Cytoplasm, (iii) Nucleus
- d)(i) tRNA, (ii) Nucleus, (iii) Cytoplasm
Correct answer is option 'A'. Can you explain this answer?
The fully processed hnRNA is called (i) and is transported out of the (ii) into (iii) for translation.
a)
(i) mRNA, (ii) Nucleus, (iii) Cytoplasm
b)
(i) mRNA, (ii) Cytoplasm, (iii) Nucleus
c)
(i) tRNA, (ii) Cytoplasm, (iii) Nucleus
d)
(i) tRNA, (ii) Nucleus, (iii) Cytoplasm
|
|
Preeti Iyer answered |
Heterogeneous nuclear RNA or hnRNA is a term that encompasses various types and sizes of RNAs found in the eukaryotic cell nucleus. When this undergoes modifications and processing it will give rise to the mature mRNA that has the inherent code for the protein synthesis (amino acid code). The mRNA in eukaryotes is synthesized in the nucleus and hence should be transported to the cytoplasm and into the ribosome for protein synthesis by the process of translation.
If a double stranded DNA has 20% of cytosines, calculate the percent of adenine in the DNA.- a)20%
- b)40%
- c)30%
- d)60%
Correct answer is option 'C'. Can you explain this answer?
If a double stranded DNA has 20% of cytosines, calculate the percent of adenine in the DNA.
a)
20%
b)
40%
c)
30%
d)
60%
|
|
Suresh Iyer answered |
In a DNA molecule, the number of cytosine molecules is equal to guanine molecules and the number of adenine molecules are equal to Thymine molecules. Thus, If a double stranded DNA has 20% of cytosine, it has 20% of guanine. Thus, cytosine plus guanine make 40% of the total bases. The remaining 60% includes both adenine and thymine which are in equal amounts. Therefore, the percent of adenine is 30%.
Given below are the steps of protein synthesis. Arrange them in. correct sequence and select the correct option.
(i) Codon-anticodon reaction between mRNA and aminoacyl tRNA complex.
(ii) Attachment of mRNA and smaller sub-unit of ribosome.
(iii) Charging or aminoacylation of tRNA.
(iv) Attachment of larger sub unit of ribosome to the mRNA-tRNAMet complex.
(v) Linking of adjacent amino acids.
(vi) Formation of polypeptide chain.- a)(ii) → (i) → (iii) → (v) → (iv) → (vi)
- b)(v) → (ii) → (i) → (iii) → (iv) → (vi)
- c)(iii) → (ii) → (iv) → (i) → (v) → (vi)
- d)(iii) → (ii) → (i) → (iv) → (v) → (vi)
Correct answer is option 'D'. Can you explain this answer?
Given below are the steps of protein synthesis. Arrange them in. correct sequence and select the correct option.
(i) Codon-anticodon reaction between mRNA and aminoacyl tRNA complex.
(ii) Attachment of mRNA and smaller sub-unit of ribosome.
(iii) Charging or aminoacylation of tRNA.
(iv) Attachment of larger sub unit of ribosome to the mRNA-tRNAMet complex.
(v) Linking of adjacent amino acids.
(vi) Formation of polypeptide chain.
(i) Codon-anticodon reaction between mRNA and aminoacyl tRNA complex.
(ii) Attachment of mRNA and smaller sub-unit of ribosome.
(iii) Charging or aminoacylation of tRNA.
(iv) Attachment of larger sub unit of ribosome to the mRNA-tRNAMet complex.
(v) Linking of adjacent amino acids.
(vi) Formation of polypeptide chain.
a)
(ii) → (i) → (iii) → (v) → (iv) → (vi)
b)
(v) → (ii) → (i) → (iii) → (iv) → (vi)
c)
(iii) → (ii) → (iv) → (i) → (v) → (vi)
d)
(iii) → (ii) → (i) → (iv) → (v) → (vi)
|
|
Anshu Kaur answered |
Overview of Protein Synthesis Steps
Protein synthesis is a vital biological process that involves several key steps. The correct sequence of events is crucial for accurate translation of mRNA into proteins.
Steps in the Correct Sequence
1. Charging or Aminoacylation of tRNA (iii)
- Before translation, tRNA molecules are charged with their respective amino acids by specific enzymes called aminoacyl-tRNA synthetases.
2. Attachment of mRNA and Smaller Sub-unit of Ribosome (ii)
- The small ribosomal subunit binds to the mRNA, scanning for the start codon (AUG), which signals the beginning of translation.
3. Attachment of Larger Sub-unit of Ribosome to the mRNA-tRNAMet Complex (iv)
- Once the start codon is recognized, the large ribosomal subunit attaches to form a complete ribosome, ready for translation.
4. Codon-Anticodon Reaction Between mRNA and Aminoacyl tRNA Complex (i)
- The charged tRNA with its corresponding amino acid binds to the mRNA codon through complementary base pairing between the codon and the tRNA's anticodon.
5. Linking of Adjacent Amino Acids (v)
- The ribosome facilitates the formation of peptide bonds between the amino acids brought by the tRNAs, elongating the polypeptide chain.
6. Formation of Polypeptide Chain (vi)
- As the ribosome moves along the mRNA, the polypeptide chain grows until a stop codon is reached, leading to the termination of translation.
Conclusion
The correct sequence (iii) → (ii) → (iv) → (i) → (v) → (vi) reflects the organized process of protein synthesis, ensuring that proteins are accurately synthesized based on the genetic code presented by mRNA. Thus, option 'D' is the correct answer.
Protein synthesis is a vital biological process that involves several key steps. The correct sequence of events is crucial for accurate translation of mRNA into proteins.
Steps in the Correct Sequence
1. Charging or Aminoacylation of tRNA (iii)
- Before translation, tRNA molecules are charged with their respective amino acids by specific enzymes called aminoacyl-tRNA synthetases.
2. Attachment of mRNA and Smaller Sub-unit of Ribosome (ii)
- The small ribosomal subunit binds to the mRNA, scanning for the start codon (AUG), which signals the beginning of translation.
3. Attachment of Larger Sub-unit of Ribosome to the mRNA-tRNAMet Complex (iv)
- Once the start codon is recognized, the large ribosomal subunit attaches to form a complete ribosome, ready for translation.
4. Codon-Anticodon Reaction Between mRNA and Aminoacyl tRNA Complex (i)
- The charged tRNA with its corresponding amino acid binds to the mRNA codon through complementary base pairing between the codon and the tRNA's anticodon.
5. Linking of Adjacent Amino Acids (v)
- The ribosome facilitates the formation of peptide bonds between the amino acids brought by the tRNAs, elongating the polypeptide chain.
6. Formation of Polypeptide Chain (vi)
- As the ribosome moves along the mRNA, the polypeptide chain grows until a stop codon is reached, leading to the termination of translation.
Conclusion
The correct sequence (iii) → (ii) → (iv) → (i) → (v) → (vi) reflects the organized process of protein synthesis, ensuring that proteins are accurately synthesized based on the genetic code presented by mRNA. Thus, option 'D' is the correct answer.
Select the incorrect statement regarding DNA replication.- a)Leading strand is formed in 5'→3' direction
- b)Okazaki fragments are formed in 5'→3' direction
- c)DNA polymerase catalyses polymerisation in 5'→3' direction
- d)DNA polymerase catalyses polymerisation in 3'→5' direction
Correct answer is option 'D'. Can you explain this answer?
Select the incorrect statement regarding DNA replication.
a)
Leading strand is formed in 5'→3' direction
b)
Okazaki fragments are formed in 5'→3' direction
c)
DNA polymerase catalyses polymerisation in 5'→3' direction
d)
DNA polymerase catalyses polymerisation in 3'→5' direction
|
|
Priya Menon answered |
Synthesis of DNA by DNA polymerases occurs only in 5'→3' direction. One strand called leading strand , is copied in the same direction as the unwinding helix. The other strand is known as lagging strand. Replication of lagging strand is in a dicontinuous way , and in the direction of growth of lagging stand is 3'→5' though in short segments of DNA which are always in the 5'→3' direction. These short segments are called Okazaki fragments joined together by the action od DNA ligase.
The structural genes, in eukaryotes possess coding and non-coding sequences called as (i) and (ii) respectively.- a)(i) promoter, (ii) operator
- b)(i) introns, (ii) exons
- c)(i) exons, (ii) introns
- d)(i) enhancer, (ii) silencer
Correct answer is option 'C'. Can you explain this answer?
The structural genes, in eukaryotes possess coding and non-coding sequences called as (i) and (ii) respectively.
a)
(i) promoter, (ii) operator
b)
(i) introns, (ii) exons
c)
(i) exons, (ii) introns
d)
(i) enhancer, (ii) silencer
|
|
Preeti Iyer answered |
In eukaryotes, the coding sequences or expressed sequences are defined as exons. Exons appear in mature or processed RNA. The exons are interrupted by introns, they do not appear in mature or processed RNA.
Which out of the following statements is incorrect?- a)Genetic code is ambiguous.
- b)Genetic code is degenerate.
- c)Genetic code is universal.
- d)Genetic code is non-overlapping.
Correct answer is option 'A'. Can you explain this answer?
Which out of the following statements is incorrect?
a)
Genetic code is ambiguous.
b)
Genetic code is degenerate.
c)
Genetic code is universal.
d)
Genetic code is non-overlapping.
|
|
Vivek Patel answered |
Since there are 64 triplet codons and only 20 amino acids, genetic code is non-ambiguous. Non-ambiguous code means that there is no ambiguity about a particular code. One codon specifies only one amino acid and not any other except GUG which normally codes for valine but in certain conditions it also codes for N-formyl methionine as initiation codon.
Q. The process of transformation is not affected by which of the following enzymes?
A. DNase
B. RNase
C. Peptidase
D. Lipase- a)A, B
- b)A, B, C, D
- c)B, C, D
- d)A, B, C
Correct answer is option 'C'. Can you explain this answer?
Q. The process of transformation is not affected by which of the following enzymes?
A. DNase
B. RNase
C. Peptidase
D. Lipase
A. DNase
B. RNase
C. Peptidase
D. Lipase
a)
A, B
b)
A, B, C, D
c)
B, C, D
d)
A, B, C
|
|
Gayatri Bose answered |
Answer:
The process of transformation is a genetic process in which foreign genetic material is introduced into a cell, resulting in a genetic change. In this process, the foreign DNA is taken up by the recipient cell and incorporated into its own genome. However, not all enzymes have an effect on the process of transformation.
Enzymes that do not affect the process of transformation:
The correct answer is option C, which includes the enzymes B. RNase, C. Peptidase, and D. Lipase. Let's understand why these enzymes do not have an effect on the process of transformation:
1. RNase:
RNase is an enzyme that degrades RNA molecules. However, in the process of transformation, the focus is on DNA molecules and their uptake by the recipient cell. Since RNase specifically targets RNA and not DNA, it does not have any effect on the transformation process.
2. Peptidase:
Peptidase is an enzyme that catalyzes the breakdown of peptides into amino acids. It is involved in protein metabolism and does not directly interact with DNA or RNA. Therefore, it does not affect the process of transformation.
3. Lipase:
Lipase is an enzyme that catalyzes the breakdown of lipids (fats). It does not have any direct involvement in DNA or RNA metabolism. Therefore, lipase has no effect on the process of transformation.
Enzymes that do affect the process of transformation:
The enzyme A. DNase, on the other hand, does have an effect on the process of transformation. DNase is an enzyme that degrades DNA molecules. If DNase is present in the environment during the transformation process, it can degrade the foreign DNA that has been taken up by the recipient cell, preventing its incorporation into the genome. Therefore, DNase can hinder the success of transformation.
To summarize, enzymes like DNase can have a negative impact on the process of transformation by degrading foreign DNA. However, enzymes like RNase, peptidase, and lipase do not affect the transformation process as they do not directly interact with DNA molecules. Therefore, the correct answer is option C.
The process of transformation is a genetic process in which foreign genetic material is introduced into a cell, resulting in a genetic change. In this process, the foreign DNA is taken up by the recipient cell and incorporated into its own genome. However, not all enzymes have an effect on the process of transformation.
Enzymes that do not affect the process of transformation:
The correct answer is option C, which includes the enzymes B. RNase, C. Peptidase, and D. Lipase. Let's understand why these enzymes do not have an effect on the process of transformation:
1. RNase:
RNase is an enzyme that degrades RNA molecules. However, in the process of transformation, the focus is on DNA molecules and their uptake by the recipient cell. Since RNase specifically targets RNA and not DNA, it does not have any effect on the transformation process.
2. Peptidase:
Peptidase is an enzyme that catalyzes the breakdown of peptides into amino acids. It is involved in protein metabolism and does not directly interact with DNA or RNA. Therefore, it does not affect the process of transformation.
3. Lipase:
Lipase is an enzyme that catalyzes the breakdown of lipids (fats). It does not have any direct involvement in DNA or RNA metabolism. Therefore, lipase has no effect on the process of transformation.
Enzymes that do affect the process of transformation:
The enzyme A. DNase, on the other hand, does have an effect on the process of transformation. DNase is an enzyme that degrades DNA molecules. If DNase is present in the environment during the transformation process, it can degrade the foreign DNA that has been taken up by the recipient cell, preventing its incorporation into the genome. Therefore, DNase can hinder the success of transformation.
To summarize, enzymes like DNase can have a negative impact on the process of transformation by degrading foreign DNA. However, enzymes like RNase, peptidase, and lipase do not affect the transformation process as they do not directly interact with DNA molecules. Therefore, the correct answer is option C.
The mutations that involve addition, deletion or substitution of a single base pair in a gene are referred to as- a)point mutations
- b)lethal mutations
- c)silent mutations
- d)retrogressive mutations
Correct answer is option 'A'. Can you explain this answer?
The mutations that involve addition, deletion or substitution of a single base pair in a gene are referred to as
a)
point mutations
b)
lethal mutations
c)
silent mutations
d)
retrogressive mutations
|
|
Suresh Iyer answered |
Most of the gene. mutations involve a change in only a single nucleotide or nitrogen base of the cistron. These gene mutations are called point mutations e.g. sickle cell anaemia in which polypeptide chain coding for haemoglobin contains valine instead of glutamic acid due to substitution of T by A in second position of triplet codon.
Which of the following pairs is incorrectly matched?- a)Purines-Adenine and Guanine
- b)Pyrimidines-Cytosine and Uracil
- c)Nucleosides-Adenosine and Thymidine
- d)DNA-Basic biomolecule
Correct answer is option 'D'. Can you explain this answer?
Which of the following pairs is incorrectly matched?
a)
Purines-Adenine and Guanine
b)
Pyrimidines-Cytosine and Uracil
c)
Nucleosides-Adenosine and Thymidine
d)
DNA-Basic biomolecule
|
|
Gaurav Kumar answered |
DNA (deoxyribonucleic acid) is an acidic biomolecule.
What is the process of activation of amino acids in the presence of ATP and its linkage to their cognate tRNA known as?- a)Aminoacetylation of ATP
- b)Charging of ATP
- c)Aminoacetylation of tRNA
- d)Charging of tRNA
Correct answer is option 'D'. Can you explain this answer?
What is the process of activation of amino acids in the presence of ATP and its linkage to their cognate tRNA known as?
a)
Aminoacetylation of ATP
b)
Charging of ATP
c)
Aminoacetylation of tRNA
d)
Charging of tRNA

|
Lead Academy answered |
- In order to form a peptide bond, a certain quantity of energy is required.
- The first phase in this process is known as charging of tRNA.
- It is also known as Aminoacylation of tRNA.
- In this process, the amino acids are activated in the presence of ATP and are linked to their cognate tRNA.
Regulation of gene expression occurs at the level of- a)transcription
- b)processing/splicing
- c)translation
- d)all of these
Correct answer is option 'D'. Can you explain this answer?
Regulation of gene expression occurs at the level of
a)
transcription
b)
processing/splicing
c)
translation
d)
all of these
|
|
Hardik Singh answered |
First of all this question's language is somewhat wrong.. there should be mentioned if they're talking about eukaryotes or prokaryotes.. For prokaryotes it's initiation transcriptional level and for eukaryotes answer would be all of these
What would happen, if in a gene encoding a polypeptide of 50 amino acids, 25th codon (UAU) is mutated to UAA?- a)A polypeptide of 49 amino acids will be formed
- b)A polypeptide of 25 amino acids will be formed
- c)A polypeptide of 24 amino acids will be formed
- d)Two polypeptides of 24 and 25 amino acids will be formed
Correct answer is option 'C'. Can you explain this answer?
What would happen, if in a gene encoding a polypeptide of 50 amino acids, 25th codon (UAU) is mutated to UAA?
a)
A polypeptide of 49 amino acids will be formed
b)
A polypeptide of 25 amino acids will be formed
c)
A polypeptide of 24 amino acids will be formed
d)
Two polypeptides of 24 and 25 amino acids will be formed
|
|
Geetika Shah answered |
UAA is a nonsense codon. It signals for polypeptide chain termination. Hence, only 24 amino acids chain will be formed.
If the sequence of bases in DNA is GCTTAGGCAA then the sequence of bases in its transcript will be- a)GCTTAGGCAA
- b)CGAATCCGTT
- c)CGAAUCCGUU
- d)AACGGAUUCG
Correct answer is option 'C'. Can you explain this answer?
If the sequence of bases in DNA is GCTTAGGCAA then the sequence of bases in its transcript will be
a)
GCTTAGGCAA
b)
CGAATCCGTT
c)
CGAAUCCGUU
d)
AACGGAUUCG
|
|
Gaurav Kumar answered |
mRNA strand is complementary to one of the DNA strands i.e., template strand. In RNA, uracil is present instead of thymine which is complementary to adenine. Cytosine and guanine are also complementary to each other. Hence, the sequence of bases in transcript would be CGAAUCCGUU.
Menthyl guanosine triphosphate is added to the 5' end of hnRNA in a process of- a)splicing
- b)capping
- c)tailing
- d)none of these
Correct answer is option 'B'. Can you explain this answer?
Menthyl guanosine triphosphate is added to the 5' end of hnRNA in a process of
a)
splicing
b)
capping
c)
tailing
d)
none of these
|
|
Diya Khanna answered |
Explanation:
Capping of hnRNA:
- The process of adding a modified nucleotide (menthyl guanosine triphosphate) to the 5' end of hnRNA is known as capping.
- This modification protects the mRNA from degradation and helps in the recognition of the mRNA by ribosomes during translation.
Function of capping:
- The capping of hnRNA is essential for the proper processing and translation of mRNA.
- It also plays a role in regulating gene expression and facilitating the export of mRNA from the nucleus to the cytoplasm.
Importance of capping:
- Capping helps in the stability of mRNA by protecting it from exonuclease degradation.
- It also assists in the initiation of translation by providing a recognition signal for ribosomes.
Therefore, in the given question, the addition of menthyl guanosine triphosphate to the 5' end of hnRNA is a part of the capping process, which is crucial for the proper functioning of mRNA in gene expression.
Capping of hnRNA:
- The process of adding a modified nucleotide (menthyl guanosine triphosphate) to the 5' end of hnRNA is known as capping.
- This modification protects the mRNA from degradation and helps in the recognition of the mRNA by ribosomes during translation.
Function of capping:
- The capping of hnRNA is essential for the proper processing and translation of mRNA.
- It also plays a role in regulating gene expression and facilitating the export of mRNA from the nucleus to the cytoplasm.
Importance of capping:
- Capping helps in the stability of mRNA by protecting it from exonuclease degradation.
- It also assists in the initiation of translation by providing a recognition signal for ribosomes.
Therefore, in the given question, the addition of menthyl guanosine triphosphate to the 5' end of hnRNA is a part of the capping process, which is crucial for the proper functioning of mRNA in gene expression.
The difference(s) between mRNA and tRNA is/are that:
(i) mRNA has more elaborate 3 - dimensional structure due to extensive base - pairing
(ii) tRNA has more elaborate 3 - dimensional structure due to extensive pairing
(iii) tRNA is usually smaller than mRNA
(iv) mRNA bears anticodon but tRNA has codons- a)(i) and (iii)
- b)All of these
- c)(ii) and (iii)
- d)(i), (ii) and (iii)
Correct answer is option 'B'. Can you explain this answer?
The difference(s) between mRNA and tRNA is/are that:
(i) mRNA has more elaborate 3 - dimensional structure due to extensive base - pairing
(ii) tRNA has more elaborate 3 - dimensional structure due to extensive pairing
(iii) tRNA is usually smaller than mRNA
(iv) mRNA bears anticodon but tRNA has codons
(i) mRNA has more elaborate 3 - dimensional structure due to extensive base - pairing
(ii) tRNA has more elaborate 3 - dimensional structure due to extensive pairing
(iii) tRNA is usually smaller than mRNA
(iv) mRNA bears anticodon but tRNA has codons
a)
(i) and (iii)
b)
All of these
c)
(ii) and (iii)
d)
(i), (ii) and (iii)
|
|
Riya Banerjee answered |
mRNA is the longest RNA with maximum molecular weight but it is least abundant. tRNA is the smallest and coiled like a clover leaf with elaborated 3-dimensional structure. mRNA consists of 75-6000 bases while tRNA has 73-93 bases.
If the sequence of bases in one strand of DNA is ATGCATGCA, what would be the sequence of bases on complementary strands?- a)ATGCATGCA
- b)AUGCAUGCA
- c)TACGTACGT
- d)UACGUACGU
Correct answer is option 'C'. Can you explain this answer?
If the sequence of bases in one strand of DNA is ATGCATGCA, what would be the sequence of bases on complementary strands?
a)
ATGCATGCA
b)
AUGCAUGCA
c)
TACGTACGT
d)
UACGUACGU
|
|
Jaspreet answered |
A complements T and C compliments G
Henceforth the complimentary DNA strand to ATGCATGCA will be "TACGTACFT" :)
Henceforth the complimentary DNA strand to ATGCATGCA will be "TACGTACFT" :)
In a mRNA molecule, untranslated regions (UTRs) are present at- a)5 - end (before start codon)
- b)3 - end (after stop codon)
- c)both (a) and (b)
- d)3 - end only
Correct answer is option 'C'. Can you explain this answer?
In a mRNA molecule, untranslated regions (UTRs) are present at
a)
5 - end (before start codon)
b)
3 - end (after stop codon)
c)
both (a) and (b)
d)
3 - end only
|
|
Harshitha Kumar answered |
In a mRNA molecule, untranslated regions (UTRs) are present at both the 5' end (before the start codon) and the 3' end (after the stop codon). The correct answer is option 'C'.
Explanation:
mRNA (messenger RNA) is a single-stranded RNA molecule that carries the genetic information from DNA to the ribosomes, where it serves as a template for protein synthesis. It contains several regions, including the coding region and the untranslated regions (UTRs).
1. Untranslated Regions (UTRs):
- Untranslated regions (UTRs) are the regions at the ends of the mRNA molecule that are not translated into protein. They are located both at the 5' end (before the start codon) and the 3' end (after the stop codon) of the mRNA molecule.
- UTRs play important regulatory roles in gene expression by influencing the stability, localization, and translation efficiency of the mRNA molecule.
- The 5' UTR, also known as the leader sequence, is present at the beginning of the mRNA molecule. It contains regulatory elements such as the Kozak sequence, which is involved in the initiation of translation.
- The 3' UTR, also known as the trailer sequence, is present at the end of the mRNA molecule. It contains regulatory elements such as binding sites for microRNAs and RNA-binding proteins, which can control mRNA stability and translation.
2. Coding Region:
- The coding region of mRNA contains the nucleotide sequence that encodes the amino acid sequence of a protein. It is located between the start codon (usually AUG) and the stop codon (UAA, UAG, or UGA).
- During translation, the ribosomes read the codons in the mRNA molecule and assemble the corresponding amino acids to form a protein.
In conclusion, untranslated regions (UTRs) are present at both the 5' and 3' ends of an mRNA molecule. They play important regulatory roles in gene expression by influencing mRNA stability, localization, and translation efficiency.
Explanation:
mRNA (messenger RNA) is a single-stranded RNA molecule that carries the genetic information from DNA to the ribosomes, where it serves as a template for protein synthesis. It contains several regions, including the coding region and the untranslated regions (UTRs).
1. Untranslated Regions (UTRs):
- Untranslated regions (UTRs) are the regions at the ends of the mRNA molecule that are not translated into protein. They are located both at the 5' end (before the start codon) and the 3' end (after the stop codon) of the mRNA molecule.
- UTRs play important regulatory roles in gene expression by influencing the stability, localization, and translation efficiency of the mRNA molecule.
- The 5' UTR, also known as the leader sequence, is present at the beginning of the mRNA molecule. It contains regulatory elements such as the Kozak sequence, which is involved in the initiation of translation.
- The 3' UTR, also known as the trailer sequence, is present at the end of the mRNA molecule. It contains regulatory elements such as binding sites for microRNAs and RNA-binding proteins, which can control mRNA stability and translation.
2. Coding Region:
- The coding region of mRNA contains the nucleotide sequence that encodes the amino acid sequence of a protein. It is located between the start codon (usually AUG) and the stop codon (UAA, UAG, or UGA).
- During translation, the ribosomes read the codons in the mRNA molecule and assemble the corresponding amino acids to form a protein.
In conclusion, untranslated regions (UTRs) are present at both the 5' and 3' ends of an mRNA molecule. They play important regulatory roles in gene expression by influencing mRNA stability, localization, and translation efficiency.
UTRs are the untranslated regions present on- a)rRNA
- b)tRNA
- c)mRNA
- d)hnRNA
Correct answer is option 'C'. Can you explain this answer?
UTRs are the untranslated regions present on
a)
rRNA
b)
tRNA
c)
mRNA
d)
hnRNA
|
|
Riya Banerjee answered |
mRNA has some additional sequences that are not translated and are referred as untranslated regions (UTRs). The UTRs are present at both the 5'-end (before the start codon) and the 3'-end (after the stop codon). They are required for an efficient translation process.
In an animal cell, the processes of transcription and translation occur in- a)nucleus only
- b)cytoplasm only
- c)nucleus and cytoplasm respectively
- d)endoplasmic reticulum
Correct answer is option 'C'. Can you explain this answer?
In an animal cell, the processes of transcription and translation occur in
a)
nucleus only
b)
cytoplasm only
c)
nucleus and cytoplasm respectively
d)
endoplasmic reticulum
|
|
Preeti Iyer answered |
As eukaryotes possess compartmentalization of cell organelles, therefore, transcription occurs in nucleus and translation occurs in the cytoplasm in eukaryotes.
Polycistronic mesenger RNA (mRNA) usually occurs in- a)bacteria
- b)prokaryotes
- c)eukaryotes
- d)both a and b
Correct answer is option 'D'. Can you explain this answer?
Polycistronic mesenger RNA (mRNA) usually occurs in
a)
bacteria
b)
prokaryotes
c)
eukaryotes
d)
both a and b
|
|
Niti Kumar answered |
Polycistronic mRNA in Bacteria and Prokaryotes
Polycistronic mRNA refers to a type of messenger RNA that carries the information for multiple genes in a single transcript. This phenomenon is commonly observed in bacteria and prokaryotes.
Bacteria
- In bacteria, the genetic material is organized in a single circular chromosome, and the transcription and translation processes are coupled.
- Bacterial mRNA is often polycistronic, meaning that a single mRNA molecule can encode multiple proteins that are part of the same functional pathway or protein complex.
- This arrangement allows for the coordinated expression of multiple genes that are involved in related cellular functions.
Prokaryotes
- Prokaryotes, including bacteria, lack a true nucleus and have a simpler organization of genetic material compared to eukaryotic cells.
- In prokaryotes, such as archaea, polycistronic mRNA is also common, serving as an efficient way to regulate gene expression and coordinate the synthesis of multiple proteins.
- The presence of polycistronic mRNA in prokaryotes allows for the rapid adaptation to changing environmental conditions and the simultaneous expression of genes that are functionally related.
Conclusion
Polycistronic mRNA is a characteristic feature of bacteria and prokaryotes, where it plays a crucial role in the regulation of gene expression and coordination of protein synthesis. This unique feature highlights the differences in gene expression mechanisms between prokaryotic and eukaryotic cells.
Polycistronic mRNA refers to a type of messenger RNA that carries the information for multiple genes in a single transcript. This phenomenon is commonly observed in bacteria and prokaryotes.
Bacteria
- In bacteria, the genetic material is organized in a single circular chromosome, and the transcription and translation processes are coupled.
- Bacterial mRNA is often polycistronic, meaning that a single mRNA molecule can encode multiple proteins that are part of the same functional pathway or protein complex.
- This arrangement allows for the coordinated expression of multiple genes that are involved in related cellular functions.
Prokaryotes
- Prokaryotes, including bacteria, lack a true nucleus and have a simpler organization of genetic material compared to eukaryotic cells.
- In prokaryotes, such as archaea, polycistronic mRNA is also common, serving as an efficient way to regulate gene expression and coordinate the synthesis of multiple proteins.
- The presence of polycistronic mRNA in prokaryotes allows for the rapid adaptation to changing environmental conditions and the simultaneous expression of genes that are functionally related.
Conclusion
Polycistronic mRNA is a characteristic feature of bacteria and prokaryotes, where it plays a crucial role in the regulation of gene expression and coordination of protein synthesis. This unique feature highlights the differences in gene expression mechanisms between prokaryotic and eukaryotic cells.
Some amino acids are coded by more than one codon, hence the genetic code is- a)overlapping
- b)degenerate
- c)wobbled
- d)unambiguous
Correct answer is option 'B'. Can you explain this answer?
Some amino acids are coded by more than one codon, hence the genetic code is
a)
overlapping
b)
degenerate
c)
wobbled
d)
unambiguous
|
|
Rishabh Chavan answered |
Explanation:
The genetic code is a set of rules by which the DNA or mRNA sequence is translated into the amino acid sequence in a protein. Each amino acid is represented by a sequence of three nucleotides called codons. However, there are more codons than there are amino acids, which means that some amino acids are coded by more than one codon. This property of the genetic code is referred to as degeneracy.
Reasons for degeneracy:
The degeneracy of the genetic code has several reasons:
1. Redundancy: There are 20 different amino acids but only four different nucleotides (A, T/U, G, C). Since each codon is composed of three nucleotides, there are a total of 64 possible codons (4^3). Therefore, multiple codons can code for the same amino acid, leading to redundancy or degeneracy.
2. Protective mechanism: Degeneracy provides a protective mechanism against mutations. Mutations in the DNA sequence can lead to changes in a codon, but if the codon still codes for the same amino acid, the resulting protein may not be functionally affected.
3. Evolutionary advantage: Degeneracy allows for more flexibility in the genetic code. It provides a buffer against errors in DNA replication or transcription, as well as allowing for adaptation to changing environmental conditions.
Examples:
Some examples of degeneracy in the genetic code include:
1. The amino acid leucine is coded by six different codons: UUA, UUG, CUU, CUC, CUA, and CUG.
2. The amino acid serine is coded by six different codons: UCU, UCC, UCA, UCG, AGU, and AGC.
3. The amino acid arginine is coded by six different codons: CGU, CGC, CGA, CGG, AGA, and AGG.
Significance:
The degeneracy of the genetic code allows for redundancy and flexibility, which is important for several biological processes:
1. Translation efficiency: Multiple codons coding for the same amino acid allow for faster and more efficient protein synthesis.
2. Evolutionary adaptation: The degeneracy of the genetic code allows for the accumulation of genetic variations over time, leading to the evolution of new traits and species.
3. Genetic engineering: The ability to use different codons to code for the same amino acid is utilized in genetic engineering techniques to optimize protein expression in various organisms.
In conclusion, the genetic code is degenerate because some amino acids are coded by more than one codon. This degeneracy provides redundancy, protection against mutations, and flexibility in the genetic code, allowing for efficient protein synthesis and evolutionary adaptation.
The genetic code is a set of rules by which the DNA or mRNA sequence is translated into the amino acid sequence in a protein. Each amino acid is represented by a sequence of three nucleotides called codons. However, there are more codons than there are amino acids, which means that some amino acids are coded by more than one codon. This property of the genetic code is referred to as degeneracy.
Reasons for degeneracy:
The degeneracy of the genetic code has several reasons:
1. Redundancy: There are 20 different amino acids but only four different nucleotides (A, T/U, G, C). Since each codon is composed of three nucleotides, there are a total of 64 possible codons (4^3). Therefore, multiple codons can code for the same amino acid, leading to redundancy or degeneracy.
2. Protective mechanism: Degeneracy provides a protective mechanism against mutations. Mutations in the DNA sequence can lead to changes in a codon, but if the codon still codes for the same amino acid, the resulting protein may not be functionally affected.
3. Evolutionary advantage: Degeneracy allows for more flexibility in the genetic code. It provides a buffer against errors in DNA replication or transcription, as well as allowing for adaptation to changing environmental conditions.
Examples:
Some examples of degeneracy in the genetic code include:
1. The amino acid leucine is coded by six different codons: UUA, UUG, CUU, CUC, CUA, and CUG.
2. The amino acid serine is coded by six different codons: UCU, UCC, UCA, UCG, AGU, and AGC.
3. The amino acid arginine is coded by six different codons: CGU, CGC, CGA, CGG, AGA, and AGG.
Significance:
The degeneracy of the genetic code allows for redundancy and flexibility, which is important for several biological processes:
1. Translation efficiency: Multiple codons coding for the same amino acid allow for faster and more efficient protein synthesis.
2. Evolutionary adaptation: The degeneracy of the genetic code allows for the accumulation of genetic variations over time, leading to the evolution of new traits and species.
3. Genetic engineering: The ability to use different codons to code for the same amino acid is utilized in genetic engineering techniques to optimize protein expression in various organisms.
In conclusion, the genetic code is degenerate because some amino acids are coded by more than one codon. This degeneracy provides redundancy, protection against mutations, and flexibility in the genetic code, allowing for efficient protein synthesis and evolutionary adaptation.
During transcription, the site of DNA molecule at which RNA polymerase binds is called- a)promoter
- b)regulator
- c)receptor
- d)enhancer
Correct answer is option 'A'. Can you explain this answer?
During transcription, the site of DNA molecule at which RNA polymerase binds is called
a)
promoter
b)
regulator
c)
receptor
d)
enhancer
|
|
Ankit Iyer answered |
Answer:
Promoter:
The site on a DNA molecule where RNA polymerase binds during transcription is called a promoter. Promoters are specific sequences of DNA that are located near the beginning of a gene.
Function of Promoter:
- Promoters play a crucial role in initiating the process of transcription by providing a binding site for RNA polymerase.
- They help in determining the starting point for transcription and also regulate the expression of genes by controlling the rate of transcription.
Types of Promoters:
- There are different types of promoters such as constitutive promoters, which are always active, and inducible promoters, which are activated in response to specific signals or environmental conditions.
Structure of Promoter:
- Promoters consist of several components, including the TATA box, which is a short sequence of DNA that helps in positioning RNA polymerase at the correct site to start transcription.
- Other elements like enhancers and silencers may also be present in the promoter region, which can influence transcriptional activity.
Importance of Promoter:
- Promoters are essential for the proper regulation of gene expression in cells. They ensure that genes are transcribed at the right time and in the right amount, which is crucial for the normal functioning of living organisms.
In conclusion, the promoter is a critical component of the transcription process as it serves as the binding site for RNA polymerase and helps in initiating the synthesis of RNA from DNA.
Promoter:
The site on a DNA molecule where RNA polymerase binds during transcription is called a promoter. Promoters are specific sequences of DNA that are located near the beginning of a gene.
Function of Promoter:
- Promoters play a crucial role in initiating the process of transcription by providing a binding site for RNA polymerase.
- They help in determining the starting point for transcription and also regulate the expression of genes by controlling the rate of transcription.
Types of Promoters:
- There are different types of promoters such as constitutive promoters, which are always active, and inducible promoters, which are activated in response to specific signals or environmental conditions.
Structure of Promoter:
- Promoters consist of several components, including the TATA box, which is a short sequence of DNA that helps in positioning RNA polymerase at the correct site to start transcription.
- Other elements like enhancers and silencers may also be present in the promoter region, which can influence transcriptional activity.
Importance of Promoter:
- Promoters are essential for the proper regulation of gene expression in cells. They ensure that genes are transcribed at the right time and in the right amount, which is crucial for the normal functioning of living organisms.
In conclusion, the promoter is a critical component of the transcription process as it serves as the binding site for RNA polymerase and helps in initiating the synthesis of RNA from DNA.
Which RNA carries the amino acids from the amino acid pool to mRNA during protein synthesis?- a)rRNA
- b)mRNA
- c)tRNA
- d)hnRNA
Correct answer is option 'C'. Can you explain this answer?
Which RNA carries the amino acids from the amino acid pool to mRNA during protein synthesis?
a)
rRNA
b)
mRNA
c)
tRNA
d)
hnRNA
|
|
Anshu Kaur answered |
Role of tRNA in Protein Synthesis
During protein synthesis, the translation process requires the transportation of amino acids to the ribosome, where they are assembled into proteins. The molecule responsible for this critical function is transfer RNA (tRNA).
What is tRNA?
- tRNA is a type of RNA that serves as an adaptor molecule in protein synthesis.
- It has a unique structure with an anticodon region that pairs with the corresponding codon on the messenger RNA (mRNA).
- Each tRNA molecule is specific to one amino acid, which it carries to the ribosome.
Function of tRNA
- Amino Acid Transport: tRNA transports amino acids from the amino acid pool in the cytoplasm to the ribosome.
- Codon Recognition: The anticodon on tRNA matches with the codon on mRNA, ensuring the correct amino acid is incorporated into the growing polypeptide chain.
- Peptide Bond Formation: Once the correct tRNA is in place at the ribosome, the amino acid it carries is added to the nascent protein via peptide bonds.
Importance in Translation
- tRNA is essential for translating the genetic code carried by mRNA into a specific sequence of amino acids.
- Without tRNA, the ribosome would not be able to correctly interpret the mRNA sequence, resulting in nonfunctional or incorrectly formed proteins.
In summary, tRNA plays a vital role in translating the genetic information from mRNA into functional proteins by transporting the necessary amino acids during protein synthesis. Thus, the correct answer to the question is option 'C' - tRNA.
During protein synthesis, the translation process requires the transportation of amino acids to the ribosome, where they are assembled into proteins. The molecule responsible for this critical function is transfer RNA (tRNA).
What is tRNA?
- tRNA is a type of RNA that serves as an adaptor molecule in protein synthesis.
- It has a unique structure with an anticodon region that pairs with the corresponding codon on the messenger RNA (mRNA).
- Each tRNA molecule is specific to one amino acid, which it carries to the ribosome.
Function of tRNA
- Amino Acid Transport: tRNA transports amino acids from the amino acid pool in the cytoplasm to the ribosome.
- Codon Recognition: The anticodon on tRNA matches with the codon on mRNA, ensuring the correct amino acid is incorporated into the growing polypeptide chain.
- Peptide Bond Formation: Once the correct tRNA is in place at the ribosome, the amino acid it carries is added to the nascent protein via peptide bonds.
Importance in Translation
- tRNA is essential for translating the genetic code carried by mRNA into a specific sequence of amino acids.
- Without tRNA, the ribosome would not be able to correctly interpret the mRNA sequence, resulting in nonfunctional or incorrectly formed proteins.
In summary, tRNA plays a vital role in translating the genetic information from mRNA into functional proteins by transporting the necessary amino acids during protein synthesis. Thus, the correct answer to the question is option 'C' - tRNA.
Other than DNA polymerase, which of the following enzymes involved in DNA synthesis?- a)Topoisomerase
- b)Helicase
- c)RNA primase
- d)All of these
Correct answer is option 'D'. Can you explain this answer?
Other than DNA polymerase, which of the following enzymes involved in DNA synthesis?
a)
Topoisomerase
b)
Helicase
c)
RNA primase
d)
All of these
|
|
Muskaan Choudhary answered |
Enzymes involved in DNA synthesis
There are several enzymes involved in DNA synthesis, apart from DNA polymerase. These enzymes play crucial roles in different steps of DNA replication. The enzymes involved in DNA synthesis include:
1. Topoisomerase:
Topoisomerase is an enzyme that helps in relieving the tension generated during DNA replication. It does so by creating temporary breaks in the DNA strands, allowing them to unwind and preventing the DNA from becoming overwound or tangled. This process is essential for the smooth movement of the replication fork and ensures accurate DNA replication.
2. Helicase:
Helicase is an enzyme responsible for unwinding the DNA double helix during replication. It uses energy from ATP hydrolysis to break the hydrogen bonds between the complementary base pairs, separating the two DNA strands. This unwinding process creates the replication fork, where DNA polymerase can then bind and synthesize new DNA strands.
3. RNA primase:
RNA primase is an enzyme that synthesizes short RNA primers that are necessary for DNA replication. These primers provide a starting point for DNA polymerase to initiate DNA synthesis. RNA primase synthesizes a short RNA segment complementary to the DNA template, which is later replaced by DNA synthesized by DNA polymerase.
Why all of these enzymes are involved in DNA synthesis?
DNA replication is a complex process that requires the coordinated action of multiple enzymes. Each enzyme performs a specific function to ensure accurate and efficient DNA synthesis. Here's why all of these enzymes are involved:
- Topoisomerase: Without topoisomerase, the tension generated during DNA unwinding would hinder the progression of the replication fork. By creating temporary breaks in the DNA strands, topoisomerase allows for smooth unwinding and prevents DNA damage.
- Helicase: Helicase plays a crucial role in unwinding the DNA double helix, separating the two strands so that DNA polymerase can synthesize new strands. It is responsible for the initiation of DNA replication and is essential for the progression of the replication fork.
- RNA primase: RNA primase synthesizes short RNA primers that are necessary for DNA replication. These primers provide a starting point for DNA polymerase to begin synthesizing the new DNA strands. Without RNA primase, DNA polymerase would not have a primer to initiate DNA synthesis.
By working together, these enzymes ensure the accurate and efficient replication of DNA, which is essential for the maintenance of genetic information and cell division.
There are several enzymes involved in DNA synthesis, apart from DNA polymerase. These enzymes play crucial roles in different steps of DNA replication. The enzymes involved in DNA synthesis include:
1. Topoisomerase:
Topoisomerase is an enzyme that helps in relieving the tension generated during DNA replication. It does so by creating temporary breaks in the DNA strands, allowing them to unwind and preventing the DNA from becoming overwound or tangled. This process is essential for the smooth movement of the replication fork and ensures accurate DNA replication.
2. Helicase:
Helicase is an enzyme responsible for unwinding the DNA double helix during replication. It uses energy from ATP hydrolysis to break the hydrogen bonds between the complementary base pairs, separating the two DNA strands. This unwinding process creates the replication fork, where DNA polymerase can then bind and synthesize new DNA strands.
3. RNA primase:
RNA primase is an enzyme that synthesizes short RNA primers that are necessary for DNA replication. These primers provide a starting point for DNA polymerase to initiate DNA synthesis. RNA primase synthesizes a short RNA segment complementary to the DNA template, which is later replaced by DNA synthesized by DNA polymerase.
Why all of these enzymes are involved in DNA synthesis?
DNA replication is a complex process that requires the coordinated action of multiple enzymes. Each enzyme performs a specific function to ensure accurate and efficient DNA synthesis. Here's why all of these enzymes are involved:
- Topoisomerase: Without topoisomerase, the tension generated during DNA unwinding would hinder the progression of the replication fork. By creating temporary breaks in the DNA strands, topoisomerase allows for smooth unwinding and prevents DNA damage.
- Helicase: Helicase plays a crucial role in unwinding the DNA double helix, separating the two strands so that DNA polymerase can synthesize new strands. It is responsible for the initiation of DNA replication and is essential for the progression of the replication fork.
- RNA primase: RNA primase synthesizes short RNA primers that are necessary for DNA replication. These primers provide a starting point for DNA polymerase to begin synthesizing the new DNA strands. Without RNA primase, DNA polymerase would not have a primer to initiate DNA synthesis.
By working together, these enzymes ensure the accurate and efficient replication of DNA, which is essential for the maintenance of genetic information and cell division.
Select the incorrectly matched pair.- a)Initiation codons - AUG, GUG
- b)Stop codons - UAA, UAG, UGA
- c)Methionine - AUG
- d)Anticodons - RNA
Correct answer is option 'D'. Can you explain this answer?
Select the incorrectly matched pair.
a)
Initiation codons - AUG, GUG
b)
Stop codons - UAA, UAG, UGA
c)
Methionine - AUG
d)
Anticodons - RNA
|
|
Lavanya Menon answered |
Polypeptide synthesis is signalled by two initiation codons - AUG (methionine) and GUG (valine). Polypeptide chain termination is signalled by three termination codons UAA (ochre), UAG (amber) and UGA (opal). They do not specify any amino acid and are hence called non-sense codons. Anticodon loop is present on tRNA.
Refer to the given sequence of steps and select the correct option.
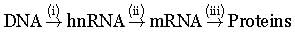
- a)(i) Replication, (ii) Transcription, (iii) - Translation
- b)(i) Replication, (ii) Processing, (iii) - Translation
- c)(i) Transcription, (ii) Splicing, (iii) - Translation
- d)(i) Transcription, (ii) Replication, (iii) - Translation
Correct answer is option 'C'. Can you explain this answer?
Refer to the given sequence of steps and select the correct option.
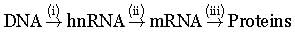
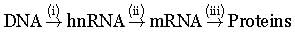
a)
(i) Replication, (ii) Transcription, (iii) - Translation
b)
(i) Replication, (ii) Processing, (iii) - Translation
c)
(i) Transcription, (ii) Splicing, (iii) - Translation
d)
(i) Transcription, (ii) Replication, (iii) - Translation
|
|
Jaspreet answered |
It's the central dogma given by Crick which depicts the unidirectional flow of information
DNA to hRNA is transcription
hRNA to mRNA is splicing in eukaryotes
mRNA to proteins is translation !!!
So correct option is "C" :)
DNA to hRNA is transcription
hRNA to mRNA is splicing in eukaryotes
mRNA to proteins is translation !!!
So correct option is "C" :)
In eukaryotes, the process of processing of primary transcript involves- a)removal of introns
- b)capping at 5' end
- c)tailing (polyadenylation) at 3' end
- d)all of these
Correct answer is option 'D'. Can you explain this answer?
In eukaryotes, the process of processing of primary transcript involves
a)
removal of introns
b)
capping at 5' end
c)
tailing (polyadenylation) at 3' end
d)
all of these
|
|
Preeti Iyer answered |
The primary mRNA transcript is longer and localised into the nucleus, where it is also called heterogeneous nuclear RNA (hnRNA) or pre-mRNA. At the 5' end of hnRNA, a cap (consisting of 7-emthyl guanosine triphosphate or 7 mG) and a tail of poly A (Adenylate residues) at the 3' end are added. These processes are respectively called as capping and tailing. The cap is a chemically modified molecule of guanosine triphosphate (GTP). The primary mRNA are made up of two types of segments, non-coding introns and the coding exons. The introns are removed by a process called RNA splicing and the exons are joined in a defined order.
Transcription unit- a)starts with TATA box
- b)starts-with pallendrous regions and ends with rho factor
- c)starts with promoter region and ends in terminator region
- d)starts with CAAT region
Correct answer is option 'C'. Can you explain this answer?
Transcription unit
a)
starts with TATA box
b)
starts-with pallendrous regions and ends with rho factor
c)
starts with promoter region and ends in terminator region
d)
starts with CAAT region
|
|
Arya Nambiar answered |
The correct answer is option 'C': Transcription starts with the promoter region and ends in the terminator region. Let's break down the process of transcription and explain why this is the correct answer.
Transcription is the first step in gene expression, where the information encoded in DNA is transcribed into RNA. This process involves several key regions and factors that play important roles. Let's discuss each option and why they are correct or incorrect.
a) Starts with TATA box:
- The TATA box is a DNA sequence found upstream of the transcription start site in many eukaryotic genes. It serves as a binding site for transcription factors and helps in the initiation of transcription.
- While the TATA box is an important element in the promoter region, it is not the starting point of transcription. Therefore, option 'a' is incorrect.
b) Starts with palindromic regions and ends with rho factor:
- Palindromic regions are DNA sequences that read the same on both strands when oriented in the same direction. They can be involved in various regulatory processes, but they are not the starting points of transcription.
- The rho factor is a protein involved in the termination of transcription in prokaryotes. It binds to the RNA molecule and causes the release of the RNA from the DNA template.
- While these regions and factors are involved in transcription, they do not mark the starting point. Therefore, option 'b' is incorrect.
c) Starts with promoter region and ends in terminator region:
- The promoter region is the DNA sequence where RNA polymerase binds and initiates transcription. It contains specific sequences and elements, such as the TATA box, that help in the recognition and binding of transcription factors.
- The terminator region is a DNA sequence that signals the end of transcription. It contains signals for the termination of RNA synthesis and the release of the RNA molecule.
- Therefore, option 'c' is correct as transcription starts with the promoter region and ends in the terminator region.
d) Starts with CAAT region:
- The CAAT box is another DNA sequence found in the promoter region of many eukaryotic genes. It is involved in the binding of transcription factors and the regulation of gene expression.
- While the CAAT region is important in the promoter, it is not the starting point of transcription. Therefore, option 'd' is incorrect.
In summary, transcription starts with the promoter region and ends in the terminator region. These regions contain specific sequences and elements that play crucial roles in the initiation and termination of transcription.
Transcription is the first step in gene expression, where the information encoded in DNA is transcribed into RNA. This process involves several key regions and factors that play important roles. Let's discuss each option and why they are correct or incorrect.
a) Starts with TATA box:
- The TATA box is a DNA sequence found upstream of the transcription start site in many eukaryotic genes. It serves as a binding site for transcription factors and helps in the initiation of transcription.
- While the TATA box is an important element in the promoter region, it is not the starting point of transcription. Therefore, option 'a' is incorrect.
b) Starts with palindromic regions and ends with rho factor:
- Palindromic regions are DNA sequences that read the same on both strands when oriented in the same direction. They can be involved in various regulatory processes, but they are not the starting points of transcription.
- The rho factor is a protein involved in the termination of transcription in prokaryotes. It binds to the RNA molecule and causes the release of the RNA from the DNA template.
- While these regions and factors are involved in transcription, they do not mark the starting point. Therefore, option 'b' is incorrect.
c) Starts with promoter region and ends in terminator region:
- The promoter region is the DNA sequence where RNA polymerase binds and initiates transcription. It contains specific sequences and elements, such as the TATA box, that help in the recognition and binding of transcription factors.
- The terminator region is a DNA sequence that signals the end of transcription. It contains signals for the termination of RNA synthesis and the release of the RNA molecule.
- Therefore, option 'c' is correct as transcription starts with the promoter region and ends in the terminator region.
d) Starts with CAAT region:
- The CAAT box is another DNA sequence found in the promoter region of many eukaryotic genes. It is involved in the binding of transcription factors and the regulation of gene expression.
- While the CAAT region is important in the promoter, it is not the starting point of transcription. Therefore, option 'd' is incorrect.
In summary, transcription starts with the promoter region and ends in the terminator region. These regions contain specific sequences and elements that play crucial roles in the initiation and termination of transcription.
Refer to the given diagram. What does it represent?
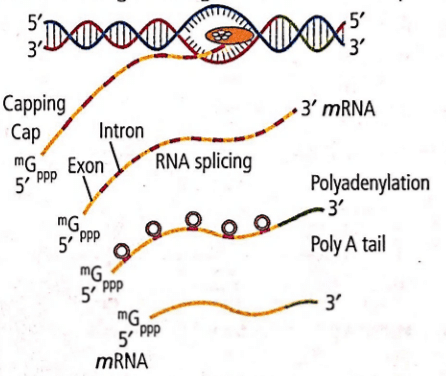
- a)Transcription in prokaryotes
- b)Transcription in eukaryotes
- c)Translation in prokaryotes
- d)Translation in eukaryotes
Correct answer is option 'B'. Can you explain this answer?
Refer to the given diagram. What does it represent?


a)
Transcription in prokaryotes
b)
Transcription in eukaryotes
c)
Translation in prokaryotes
d)
Translation in eukaryotes
|
|
Ananya Das answered |
The given diagram represents post-transcriptional processing resulting in the formation of mRNA. Since, introns and exons are present, it is transcription in eukaryotes.
Choose the correct answer from the alternatives given:
Identify the labels A, B, C and Din the given structure of tRNA and select the correct option.
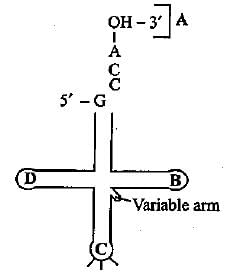
- a)A - Anticodon loop, B TΨC loop, C - AA binding site, D - DHU loop
- b)A - AA binding site, B - TΨC loop, C - Anticodon loop, D - DHU loop
- c)A - M binding site, B - DHU loop, C - Anticodon loop, D - TΨC loop
- d)A - M binding site, B - DHU loop, C - TΨC loop, D - Anticodon loop
Correct answer is option 'B'. Can you explain this answer?
Choose the correct answer from the alternatives given:
Identify the labels A, B, C and Din the given structure of tRNA and select the correct option.

Identify the labels A, B, C and Din the given structure of tRNA and select the correct option.

a)
A - Anticodon loop, B TΨC loop, C - AA binding site, D - DHU loop
b)
A - AA binding site, B - TΨC loop, C - Anticodon loop, D - DHU loop
c)
A - M binding site, B - DHU loop, C - Anticodon loop, D - TΨC loop
d)
A - M binding site, B - DHU loop, C - TΨC loop, D - Anticodon loop
|
|
Ananya Das answered |
In tRNA, there is a TWC loop which contains pseudouridine and ribothymidine. The loop is the site for attaching to ribosomes. Another loop, DHU loop contains dihydrouridine. It is a binding site for aminoacyl synthetase enzyme. tRNA molecules have unpaired (single-stranded) CCAOH sequence at the 3 end. This is called amino acid binding site because the amino acid becomes covalently attached to adenylic acid or A of CCA sequence during polypeptide synthesis. Anticodon loop is made up of three nitrogen bases for recognizing and attaching to the codon of mRNA.
Choose the correct answer from the alternatives given :
Direction : Read the sequence of nucleotides in the given segment of mRNA and the respective amino acid sequence in the polypeptide chain.
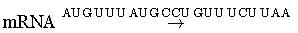
Polypeptide Met-Phe-Met-Pro-Val-Ser
Which codons respectively code for proline and valine amino acids in the given polypeptide chain, respectively?
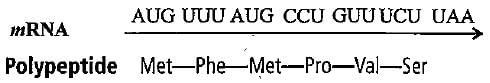
- a)CCU and GUU
- b)GUU and UCU
- c)UCU and UAA
- d)GUU and CCU
Correct answer is option 'A'. Can you explain this answer?
Choose the correct answer from the alternatives given :
Direction : Read the sequence of nucleotides in the given segment of mRNA and the respective amino acid sequence in the polypeptide chain.
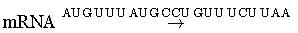
Polypeptide Met-Phe-Met-Pro-Val-Ser
Which codons respectively code for proline and valine amino acids in the given polypeptide chain, respectively?
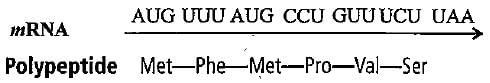
Direction : Read the sequence of nucleotides in the given segment of mRNA and the respective amino acid sequence in the polypeptide chain.
Polypeptide Met-Phe-Met-Pro-Val-Ser
Which codons respectively code for proline and valine amino acids in the given polypeptide chain, respectively?
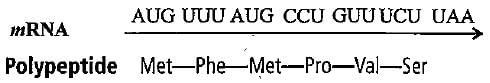
a)
CCU and GUU
b)
GUU and UCU
c)
UCU and UAA
d)
GUU and CCU
|
|
Jyoti Sengupta answered |
codon is a sequence of three nucleotides on an mRNA strand that encodes a specific amino acid.
Sixty-four different three nucleotide combinations (codons) can be made using the four nucleotides in mRNA (43 = 64 combinations).
Following are the codons that represent Valine and proline respectively.
Valine: GUA, GUC, GUG, GUU
proline: CCT, CCC, CCA, CCG
Sixty-four different three nucleotide combinations (codons) can be made using the four nucleotides in mRNA (43 = 64 combinations).
Following are the codons that represent Valine and proline respectively.
Valine: GUA, GUC, GUG, GUU
proline: CCT, CCC, CCA, CCG
During translation, activated amino acids get linked to (RNA). This process is commonly called as- a)charging of tRNA
- b)discharging of tRNA
- c)aminoacylation of tRNA
- d)both (a) and (c)
Correct answer is option 'D'. Can you explain this answer?
During translation, activated amino acids get linked to (RNA). This process is commonly called as
a)
charging of tRNA
b)
discharging of tRNA
c)
aminoacylation of tRNA
d)
both (a) and (c)
|
|
Geetika Shah answered |
During translation, a (RNA) is specifically linked to an amino acid, this process takes place under the direction of an enzyme, aminoacyl tRNA synthetase that is extremely specific, i.e., recognises only one amino acid. (RNA) complexed with amino acid is called as charged tRNA. The process is referred to a charging or aminoacylation of tRNA.
Choose the correct answer from the alternatives given:
Which of the following criteria should be fulfilled by a molecule to act as a genetic material?
(i) It should be able to replicate.
(ii) It should be structurally and chemically stable.
(iii) It should be able to undergo slow mutations.
(iv) It should be able to express itself in the form of Mendelian characters.- a)(i) and (ii)
- b)(ii) and (iii)
- c)(i), (ii) and (iii)
- d)(i), (ii), (iii) and (iv)
Correct answer is option 'D'. Can you explain this answer?
Choose the correct answer from the alternatives given:
Which of the following criteria should be fulfilled by a molecule to act as a genetic material?
(i) It should be able to replicate.
(ii) It should be structurally and chemically stable.
(iii) It should be able to undergo slow mutations.
(iv) It should be able to express itself in the form of Mendelian characters.
Which of the following criteria should be fulfilled by a molecule to act as a genetic material?
(i) It should be able to replicate.
(ii) It should be structurally and chemically stable.
(iii) It should be able to undergo slow mutations.
(iv) It should be able to express itself in the form of Mendelian characters.
a)
(i) and (ii)
b)
(ii) and (iii)
c)
(i), (ii) and (iii)
d)
(i), (ii), (iii) and (iv)
|
|
Jaideep Chauhan answered |
Criteria for a molecule to act as a genetic material
A molecule that acts as genetic material must fulfill certain criteria to ensure the proper transmission of genetic information from one generation to the next. The criteria are:
1. It should be able to replicate:
The molecule must be able to replicate itself accurately and efficiently so that the genetic information can be passed on to the next generation without any errors.
2. It should be structurally and chemically stable:
The molecule must be chemically and structurally stable so that it can withstand the various chemical and physical processes that occur during replication and cell division.
3. It should be able to undergo slow mutations:
The molecule must be able to undergo slow mutations, which are necessary for the generation of genetic diversity and evolution.
4. It should be able to express itself in the form of Mendelian characters:
The molecule must be able to express itself in the form of Mendelian characters, which are the observable traits that are inherited from one generation to the next.
Correct answer:
The correct answer is option D, which includes all the above criteria. A molecule that acts as genetic material must be able to replicate accurately, be structurally and chemically stable, undergo slow mutations, and express itself in the form of Mendelian characters.
A molecule that acts as genetic material must fulfill certain criteria to ensure the proper transmission of genetic information from one generation to the next. The criteria are:
1. It should be able to replicate:
The molecule must be able to replicate itself accurately and efficiently so that the genetic information can be passed on to the next generation without any errors.
2. It should be structurally and chemically stable:
The molecule must be chemically and structurally stable so that it can withstand the various chemical and physical processes that occur during replication and cell division.
3. It should be able to undergo slow mutations:
The molecule must be able to undergo slow mutations, which are necessary for the generation of genetic diversity and evolution.
4. It should be able to express itself in the form of Mendelian characters:
The molecule must be able to express itself in the form of Mendelian characters, which are the observable traits that are inherited from one generation to the next.
Correct answer:
The correct answer is option D, which includes all the above criteria. A molecule that acts as genetic material must be able to replicate accurately, be structurally and chemically stable, undergo slow mutations, and express itself in the form of Mendelian characters.
DNA replication takes place at ________ phase of the cell cycle- a)G1
- b)S
- c)G2
- d)M
Correct answer is option 'B'. Can you explain this answer?
DNA replication takes place at ________ phase of the cell cycle
a)
G1
b)
S
c)
G2
d)
M
|
|
Meera Singh answered |
In eukaryotes, the replication of DNA takes place at S-phase of the cell cycle. The replication of DNA and cell division cycle should be coordinated. A failure in cell division after DNA replication results into chromosomal anomaly.
Choose the correct answer from the alternatives given :
Match column I with column II and select the correct option from the given codes.
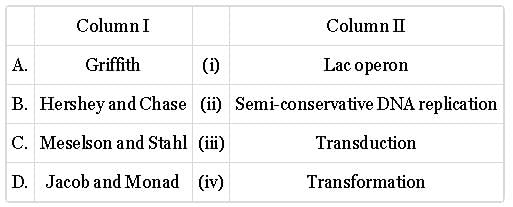
- a)A - (iv), B - (iii), C - (ii),D - (i)
- b)A - (iii), B-(iv), C - (ii),D - (i)
- c)A - (iv), B - (ii), C - (iii),D - (i)
- d)A - (ii), B - (i), C - (iii), D - (iv)
Correct answer is option 'A'. Can you explain this answer?
Choose the correct answer from the alternatives given :
Match column I with column II and select the correct option from the given codes.

Match column I with column II and select the correct option from the given codes.

a)
A - (iv), B - (iii), C - (ii),D - (i)
b)
A - (iii), B-(iv), C - (ii),D - (i)
c)
A - (iv), B - (ii), C - (iii),D - (i)
d)
A - (ii), B - (i), C - (iii), D - (iv)
|
|
Anjali Sharma answered |
Griffith performed the transformation experiment on S.pneumoniae to prove that the genetic material was inheritable. Hershey and Chase performed the transduction experiment which is the infection of a bacteria by a virus called bacteriophage to prove that DNA and not proteins were the genetic material. Meselson and Stahl the experiment on E.coli to establish that replication was semi-conservative (half of parent and half synthesized). Jacob and Monad elucidated the Lac operon a transcription-ally regulated system.
Match column I with column II and select the correct option from the given codes.
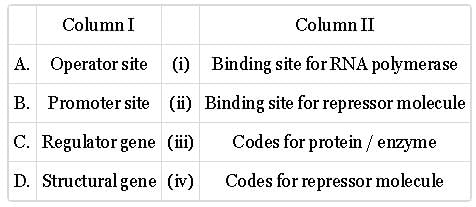
- a)A - (ii), B - (I), C - (iii), D - (iv)
- b)A - (ii), B - (i), C - (iv), D - (iii)
- c)A - (iv), B - (iii), C - (i), D - (ii)
- d)A- (ii), B - (iii), C (i), D - (iv)
Correct answer is option 'B'. Can you explain this answer?
Match column I with column II and select the correct option from the given codes.


a)
A - (ii), B - (I), C - (iii), D - (iv)
b)
A - (ii), B - (i), C - (iv), D - (iii)
c)
A - (iv), B - (iii), C - (i), D - (ii)
d)
A- (ii), B - (iii), C (i), D - (iv)
|
|
Geetika Shah answered |
The accessibility of promoter regions of prokaryotic DNA is regulated by interaction of proteins with sequences called as operators. Repressor molecules that repress gene expression bind to the operator sits and as a result the promoter cannot initiate gene transcription. Promoter is the site for the binding of the RNA polymerase to initiate the process of transcription. It refers to a conserved sequence that is specific for the binding of the RNA polymerase that can bring the expression genes for various functions. Regulator gene is the gene of an operon that codes for a repressor molecule that can bind to the operator sequence and stop gene expression. Structural gene codes for proteins or enzymes that are essential for the cell growth and survival
So, the correct option is 'A - (ii), B - (i), C - (iv), D - (iii)'.
So, the correct option is 'A - (ii), B - (i), C - (iv), D - (iii)'.
Chapter doubts & questions for Molecular Basis of Inheritance - Biology Class 12 2025 is part of NEET exam preparation. The chapters have been prepared according to the NEET exam syllabus. The Chapter doubts & questions, notes, tests & MCQs are made for NEET 2025 Exam. Find important definitions, questions, notes, meanings, examples, exercises, MCQs and online tests here.
Chapter doubts & questions of Molecular Basis of Inheritance - Biology Class 12 in English & Hindi are available as part of NEET exam.
Download more important topics, notes, lectures and mock test series for NEET Exam by signing up for free.

Contact Support
Our team is online on weekdays between 10 AM - 7 PM
Typical reply within 3 hours
|
Free Exam Preparation
at your Fingertips!
Access Free Study Material - Test Series, Structured Courses, Free Videos & Study Notes and Prepare for Your Exam With Ease

 Join the 10M+ students on EduRev
Join the 10M+ students on EduRev
|

|
Create your account for free
OR
Forgot Password
OR
Signup to see your scores
go up within 7 days!
Access 1000+ FREE Docs, Videos and Tests
Takes less than 10 seconds to signup










