Software Development Exam > Software Development Questions > Catalase and other PTS1-bearing proteins, suc...
Start Learning for Free
Catalase and other PTS1-bearing proteins, such as PMP70, are imported into the peroxisomal matrix commencing in the cytosol. For the correct insertion of proteins like PMP70 into the developing peroxisomal membrane, a complex of Pex3 and Pex16 is necessary. First, PTS1 interacts to the Pex5 receptor that has the extraordinary ability to transform in a combination with the membrane protein Pex14 from a monomeric soluble form to an oligomeric form entrenched in the peroxisomal membrane. The matrix protein separates from Pex5 and is released into the peroxisomal matrix following the formation of the matrix protein-Pex5-Pex14 complex. The membrane proteins Pex2, Pex10, and Pex1 then ubiquitinylate Pex5, which causes it to be returned to the cytoplasm. Cells from Pex12 and Pex3 deficient patients were stained with fluorescent antibodies to PMP70 or with fluorescent antibodies to catalase, a peroxisomal matrix protein, then viewed in a fluorescence microscope. What are observations possible?
- a)In wild-type cells, peroxisomal membrane and matrix proteins are NOT visible as bright foci in numerous peroxisomal bodies.
- b)In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is localized normally to peroxisomal bodies
- c)In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is NOT localized normally to peroxisomal bodies
- d)In cells from a Pex3-deficient patient, peroxisomal membranes CAN assemble, and as a consequence, peroxisomal bodies do not form. Thus both catalase and PMP70 are mis-localized to the cytosol.
Correct answer is option 'B'. Can you explain this answer?
Verified Answer
Catalase and other PTS1-bearing proteins, such as PMP70, are imported ...
Concept:
- Peroxisomes are small organelles bounded by a single membrane.
- Unlike mitochondria and chloroplasts, peroxisomes lack DNA and ribosomes.
- Thus all luminal peroxisomal proteins are encoded by nuclear genes, synthesized on free ribosomes in the cytosol, and then incorporated into preexisting or newly generated peroxisomes.
Explanation:
Fig 1: PTS1-directed import of peroxisomal matrix proteins
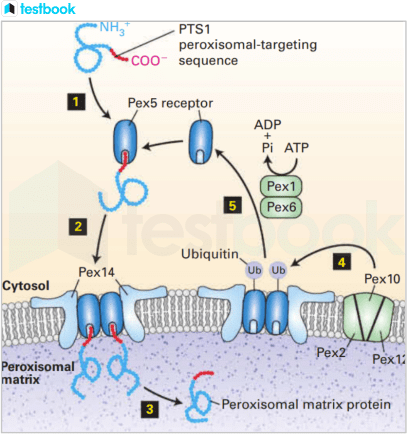

- Most peroxisomal matrix proteins contain a C- terminal PTS1 targeting sequence (red), which binds to the cytosolic receptor Pex5.
Step 2 : Pex5 with the bound matrix protein forms a multimeric complex with the Pex14 receptor located on the peroxisomal membrane.
Step 3 : After assembly of the matrix protein-Pex5-Pex14 complex, the matrix protein dissociates from Pex5 and is released into the peroxisomal matrix.
Steps 4 and 5 : Pex5 is then returned to the cytosol by a process that involves ubiquitinylation by the membrane proteins Pex2, Pex10, and Pex12, followed by ATP-dependent removal from the membrane by the AAA-ATPase proteins Pex1 and Pex6.
Option 1:
In wild-type cells, peroxisomal membrane and matrix proteins are NOT visible as bright foci in numerous peroxisomal bodies.
- As Cells were stained with fluorescent antibodies to PMP70, a peroxisomal membrane protein, or with fluorescent antibodies to catalase, they should be visible in normal cells
- thus this option is not true
Option 2:
In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is localized normally to peroxisomal bodies
- Catalase and other PTS1-bearing proteins, such as PMP70, are imported into the peroxisomal matrix commencing in the cytosol, so in Pex12-deficient patient catalses can be present but PMP70 is PTS1-bearing proteins and does notneeds functional Pex 12 for transport.
- thus this option is true
Option 3:
In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is NOT localized normally to peroxisomal bodies
- Consider the explanation above thus this option is not true
Option 4:
In cells from a Pex3-deficient patient, peroxisomal membranes cannot assemble, and as a consequence, peroxisomal bodies do not form. Thus both catalase and PMP70 are mis-localized to the cytosol.
- Consider the query it is clear that Pex-3 is important for peroxisomes synthesis
- thus this option is not true
hence the correct answer is option 2

|
Explore Courses for Software Development exam
|

|
Similar Software Development Doubts
Catalase and other PTS1-bearing proteins, such as PMP70, are imported into the peroxisomal matrix commencing in the cytosol. For the correct insertion of proteins like PMP70 into the developing peroxisomal membrane, a complex of Pex3 and Pex16 is necessary. First, PTS1 interacts to the Pex5 receptor that has the extraordinary ability to transform in a combination with the membrane protein Pex14 from a monomeric soluble form to an oligomeric form entrenched in the peroxisomal membrane. The matrix protein separates from Pex5 and is released into the peroxisomal matrix following the formation of the matrix protein-Pex5-Pex14 complex. The membrane proteins Pex2, Pex10, and Pex1 then ubiquitinylate Pex5, which causes it to be returned to the cytoplasm.Cells from Pex12 and Pex3 deficient patients were stained with fluorescent antibodies to PMP70 or with fluorescent antibodies to catalase, a peroxisomal matrix protein, then viewed in a fluorescence microscope. What are observations possible?a)In wild-type cells, peroxisomal membrane and matrix proteins are NOT visible as bright foci in numerous peroxisomal bodies.b)In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is localized normally to peroxisomal bodiesc)In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is NOT localized normally to peroxisomal bodiesd)In cells from a Pex3-deficient patient, peroxisomal membranes CANassemble, and as a consequence, peroxisomal bodies do not form. Thus both catalase and PMP70 are mis-localized to the cytosol.Correct answer is option 'B'. Can you explain this answer?
Question Description
Catalase and other PTS1-bearing proteins, such as PMP70, are imported into the peroxisomal matrix commencing in the cytosol. For the correct insertion of proteins like PMP70 into the developing peroxisomal membrane, a complex of Pex3 and Pex16 is necessary. First, PTS1 interacts to the Pex5 receptor that has the extraordinary ability to transform in a combination with the membrane protein Pex14 from a monomeric soluble form to an oligomeric form entrenched in the peroxisomal membrane. The matrix protein separates from Pex5 and is released into the peroxisomal matrix following the formation of the matrix protein-Pex5-Pex14 complex. The membrane proteins Pex2, Pex10, and Pex1 then ubiquitinylate Pex5, which causes it to be returned to the cytoplasm.Cells from Pex12 and Pex3 deficient patients were stained with fluorescent antibodies to PMP70 or with fluorescent antibodies to catalase, a peroxisomal matrix protein, then viewed in a fluorescence microscope. What are observations possible?a)In wild-type cells, peroxisomal membrane and matrix proteins are NOT visible as bright foci in numerous peroxisomal bodies.b)In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is localized normally to peroxisomal bodiesc)In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is NOT localized normally to peroxisomal bodiesd)In cells from a Pex3-deficient patient, peroxisomal membranes CANassemble, and as a consequence, peroxisomal bodies do not form. Thus both catalase and PMP70 are mis-localized to the cytosol.Correct answer is option 'B'. Can you explain this answer? for Software Development 2025 is part of Software Development preparation. The Question and answers have been prepared according to the Software Development exam syllabus. Information about Catalase and other PTS1-bearing proteins, such as PMP70, are imported into the peroxisomal matrix commencing in the cytosol. For the correct insertion of proteins like PMP70 into the developing peroxisomal membrane, a complex of Pex3 and Pex16 is necessary. First, PTS1 interacts to the Pex5 receptor that has the extraordinary ability to transform in a combination with the membrane protein Pex14 from a monomeric soluble form to an oligomeric form entrenched in the peroxisomal membrane. The matrix protein separates from Pex5 and is released into the peroxisomal matrix following the formation of the matrix protein-Pex5-Pex14 complex. The membrane proteins Pex2, Pex10, and Pex1 then ubiquitinylate Pex5, which causes it to be returned to the cytoplasm.Cells from Pex12 and Pex3 deficient patients were stained with fluorescent antibodies to PMP70 or with fluorescent antibodies to catalase, a peroxisomal matrix protein, then viewed in a fluorescence microscope. What are observations possible?a)In wild-type cells, peroxisomal membrane and matrix proteins are NOT visible as bright foci in numerous peroxisomal bodies.b)In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is localized normally to peroxisomal bodiesc)In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is NOT localized normally to peroxisomal bodiesd)In cells from a Pex3-deficient patient, peroxisomal membranes CANassemble, and as a consequence, peroxisomal bodies do not form. Thus both catalase and PMP70 are mis-localized to the cytosol.Correct answer is option 'B'. Can you explain this answer? covers all topics & solutions for Software Development 2025 Exam. Find important definitions, questions, meanings, examples, exercises and tests below for Catalase and other PTS1-bearing proteins, such as PMP70, are imported into the peroxisomal matrix commencing in the cytosol. For the correct insertion of proteins like PMP70 into the developing peroxisomal membrane, a complex of Pex3 and Pex16 is necessary. First, PTS1 interacts to the Pex5 receptor that has the extraordinary ability to transform in a combination with the membrane protein Pex14 from a monomeric soluble form to an oligomeric form entrenched in the peroxisomal membrane. The matrix protein separates from Pex5 and is released into the peroxisomal matrix following the formation of the matrix protein-Pex5-Pex14 complex. The membrane proteins Pex2, Pex10, and Pex1 then ubiquitinylate Pex5, which causes it to be returned to the cytoplasm.Cells from Pex12 and Pex3 deficient patients were stained with fluorescent antibodies to PMP70 or with fluorescent antibodies to catalase, a peroxisomal matrix protein, then viewed in a fluorescence microscope. What are observations possible?a)In wild-type cells, peroxisomal membrane and matrix proteins are NOT visible as bright foci in numerous peroxisomal bodies.b)In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is localized normally to peroxisomal bodiesc)In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is NOT localized normally to peroxisomal bodiesd)In cells from a Pex3-deficient patient, peroxisomal membranes CANassemble, and as a consequence, peroxisomal bodies do not form. Thus both catalase and PMP70 are mis-localized to the cytosol.Correct answer is option 'B'. Can you explain this answer?.
Catalase and other PTS1-bearing proteins, such as PMP70, are imported into the peroxisomal matrix commencing in the cytosol. For the correct insertion of proteins like PMP70 into the developing peroxisomal membrane, a complex of Pex3 and Pex16 is necessary. First, PTS1 interacts to the Pex5 receptor that has the extraordinary ability to transform in a combination with the membrane protein Pex14 from a monomeric soluble form to an oligomeric form entrenched in the peroxisomal membrane. The matrix protein separates from Pex5 and is released into the peroxisomal matrix following the formation of the matrix protein-Pex5-Pex14 complex. The membrane proteins Pex2, Pex10, and Pex1 then ubiquitinylate Pex5, which causes it to be returned to the cytoplasm.Cells from Pex12 and Pex3 deficient patients were stained with fluorescent antibodies to PMP70 or with fluorescent antibodies to catalase, a peroxisomal matrix protein, then viewed in a fluorescence microscope. What are observations possible?a)In wild-type cells, peroxisomal membrane and matrix proteins are NOT visible as bright foci in numerous peroxisomal bodies.b)In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is localized normally to peroxisomal bodiesc)In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is NOT localized normally to peroxisomal bodiesd)In cells from a Pex3-deficient patient, peroxisomal membranes CANassemble, and as a consequence, peroxisomal bodies do not form. Thus both catalase and PMP70 are mis-localized to the cytosol.Correct answer is option 'B'. Can you explain this answer? for Software Development 2025 is part of Software Development preparation. The Question and answers have been prepared according to the Software Development exam syllabus. Information about Catalase and other PTS1-bearing proteins, such as PMP70, are imported into the peroxisomal matrix commencing in the cytosol. For the correct insertion of proteins like PMP70 into the developing peroxisomal membrane, a complex of Pex3 and Pex16 is necessary. First, PTS1 interacts to the Pex5 receptor that has the extraordinary ability to transform in a combination with the membrane protein Pex14 from a monomeric soluble form to an oligomeric form entrenched in the peroxisomal membrane. The matrix protein separates from Pex5 and is released into the peroxisomal matrix following the formation of the matrix protein-Pex5-Pex14 complex. The membrane proteins Pex2, Pex10, and Pex1 then ubiquitinylate Pex5, which causes it to be returned to the cytoplasm.Cells from Pex12 and Pex3 deficient patients were stained with fluorescent antibodies to PMP70 or with fluorescent antibodies to catalase, a peroxisomal matrix protein, then viewed in a fluorescence microscope. What are observations possible?a)In wild-type cells, peroxisomal membrane and matrix proteins are NOT visible as bright foci in numerous peroxisomal bodies.b)In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is localized normally to peroxisomal bodiesc)In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is NOT localized normally to peroxisomal bodiesd)In cells from a Pex3-deficient patient, peroxisomal membranes CANassemble, and as a consequence, peroxisomal bodies do not form. Thus both catalase and PMP70 are mis-localized to the cytosol.Correct answer is option 'B'. Can you explain this answer? covers all topics & solutions for Software Development 2025 Exam. Find important definitions, questions, meanings, examples, exercises and tests below for Catalase and other PTS1-bearing proteins, such as PMP70, are imported into the peroxisomal matrix commencing in the cytosol. For the correct insertion of proteins like PMP70 into the developing peroxisomal membrane, a complex of Pex3 and Pex16 is necessary. First, PTS1 interacts to the Pex5 receptor that has the extraordinary ability to transform in a combination with the membrane protein Pex14 from a monomeric soluble form to an oligomeric form entrenched in the peroxisomal membrane. The matrix protein separates from Pex5 and is released into the peroxisomal matrix following the formation of the matrix protein-Pex5-Pex14 complex. The membrane proteins Pex2, Pex10, and Pex1 then ubiquitinylate Pex5, which causes it to be returned to the cytoplasm.Cells from Pex12 and Pex3 deficient patients were stained with fluorescent antibodies to PMP70 or with fluorescent antibodies to catalase, a peroxisomal matrix protein, then viewed in a fluorescence microscope. What are observations possible?a)In wild-type cells, peroxisomal membrane and matrix proteins are NOT visible as bright foci in numerous peroxisomal bodies.b)In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is localized normally to peroxisomal bodiesc)In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is NOT localized normally to peroxisomal bodiesd)In cells from a Pex3-deficient patient, peroxisomal membranes CANassemble, and as a consequence, peroxisomal bodies do not form. Thus both catalase and PMP70 are mis-localized to the cytosol.Correct answer is option 'B'. Can you explain this answer?.
Solutions for Catalase and other PTS1-bearing proteins, such as PMP70, are imported into the peroxisomal matrix commencing in the cytosol. For the correct insertion of proteins like PMP70 into the developing peroxisomal membrane, a complex of Pex3 and Pex16 is necessary. First, PTS1 interacts to the Pex5 receptor that has the extraordinary ability to transform in a combination with the membrane protein Pex14 from a monomeric soluble form to an oligomeric form entrenched in the peroxisomal membrane. The matrix protein separates from Pex5 and is released into the peroxisomal matrix following the formation of the matrix protein-Pex5-Pex14 complex. The membrane proteins Pex2, Pex10, and Pex1 then ubiquitinylate Pex5, which causes it to be returned to the cytoplasm.Cells from Pex12 and Pex3 deficient patients were stained with fluorescent antibodies to PMP70 or with fluorescent antibodies to catalase, a peroxisomal matrix protein, then viewed in a fluorescence microscope. What are observations possible?a)In wild-type cells, peroxisomal membrane and matrix proteins are NOT visible as bright foci in numerous peroxisomal bodies.b)In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is localized normally to peroxisomal bodiesc)In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is NOT localized normally to peroxisomal bodiesd)In cells from a Pex3-deficient patient, peroxisomal membranes CANassemble, and as a consequence, peroxisomal bodies do not form. Thus both catalase and PMP70 are mis-localized to the cytosol.Correct answer is option 'B'. Can you explain this answer? in English & in Hindi are available as part of our courses for Software Development.
Download more important topics, notes, lectures and mock test series for Software Development Exam by signing up for free.
Here you can find the meaning of Catalase and other PTS1-bearing proteins, such as PMP70, are imported into the peroxisomal matrix commencing in the cytosol. For the correct insertion of proteins like PMP70 into the developing peroxisomal membrane, a complex of Pex3 and Pex16 is necessary. First, PTS1 interacts to the Pex5 receptor that has the extraordinary ability to transform in a combination with the membrane protein Pex14 from a monomeric soluble form to an oligomeric form entrenched in the peroxisomal membrane. The matrix protein separates from Pex5 and is released into the peroxisomal matrix following the formation of the matrix protein-Pex5-Pex14 complex. The membrane proteins Pex2, Pex10, and Pex1 then ubiquitinylate Pex5, which causes it to be returned to the cytoplasm.Cells from Pex12 and Pex3 deficient patients were stained with fluorescent antibodies to PMP70 or with fluorescent antibodies to catalase, a peroxisomal matrix protein, then viewed in a fluorescence microscope. What are observations possible?a)In wild-type cells, peroxisomal membrane and matrix proteins are NOT visible as bright foci in numerous peroxisomal bodies.b)In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is localized normally to peroxisomal bodiesc)In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is NOT localized normally to peroxisomal bodiesd)In cells from a Pex3-deficient patient, peroxisomal membranes CANassemble, and as a consequence, peroxisomal bodies do not form. Thus both catalase and PMP70 are mis-localized to the cytosol.Correct answer is option 'B'. Can you explain this answer? defined & explained in the simplest way possible. Besides giving the explanation of
Catalase and other PTS1-bearing proteins, such as PMP70, are imported into the peroxisomal matrix commencing in the cytosol. For the correct insertion of proteins like PMP70 into the developing peroxisomal membrane, a complex of Pex3 and Pex16 is necessary. First, PTS1 interacts to the Pex5 receptor that has the extraordinary ability to transform in a combination with the membrane protein Pex14 from a monomeric soluble form to an oligomeric form entrenched in the peroxisomal membrane. The matrix protein separates from Pex5 and is released into the peroxisomal matrix following the formation of the matrix protein-Pex5-Pex14 complex. The membrane proteins Pex2, Pex10, and Pex1 then ubiquitinylate Pex5, which causes it to be returned to the cytoplasm.Cells from Pex12 and Pex3 deficient patients were stained with fluorescent antibodies to PMP70 or with fluorescent antibodies to catalase, a peroxisomal matrix protein, then viewed in a fluorescence microscope. What are observations possible?a)In wild-type cells, peroxisomal membrane and matrix proteins are NOT visible as bright foci in numerous peroxisomal bodies.b)In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is localized normally to peroxisomal bodiesc)In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is NOT localized normally to peroxisomal bodiesd)In cells from a Pex3-deficient patient, peroxisomal membranes CANassemble, and as a consequence, peroxisomal bodies do not form. Thus both catalase and PMP70 are mis-localized to the cytosol.Correct answer is option 'B'. Can you explain this answer?, a detailed solution for Catalase and other PTS1-bearing proteins, such as PMP70, are imported into the peroxisomal matrix commencing in the cytosol. For the correct insertion of proteins like PMP70 into the developing peroxisomal membrane, a complex of Pex3 and Pex16 is necessary. First, PTS1 interacts to the Pex5 receptor that has the extraordinary ability to transform in a combination with the membrane protein Pex14 from a monomeric soluble form to an oligomeric form entrenched in the peroxisomal membrane. The matrix protein separates from Pex5 and is released into the peroxisomal matrix following the formation of the matrix protein-Pex5-Pex14 complex. The membrane proteins Pex2, Pex10, and Pex1 then ubiquitinylate Pex5, which causes it to be returned to the cytoplasm.Cells from Pex12 and Pex3 deficient patients were stained with fluorescent antibodies to PMP70 or with fluorescent antibodies to catalase, a peroxisomal matrix protein, then viewed in a fluorescence microscope. What are observations possible?a)In wild-type cells, peroxisomal membrane and matrix proteins are NOT visible as bright foci in numerous peroxisomal bodies.b)In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is localized normally to peroxisomal bodiesc)In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is NOT localized normally to peroxisomal bodiesd)In cells from a Pex3-deficient patient, peroxisomal membranes CANassemble, and as a consequence, peroxisomal bodies do not form. Thus both catalase and PMP70 are mis-localized to the cytosol.Correct answer is option 'B'. Can you explain this answer? has been provided alongside types of Catalase and other PTS1-bearing proteins, such as PMP70, are imported into the peroxisomal matrix commencing in the cytosol. For the correct insertion of proteins like PMP70 into the developing peroxisomal membrane, a complex of Pex3 and Pex16 is necessary. First, PTS1 interacts to the Pex5 receptor that has the extraordinary ability to transform in a combination with the membrane protein Pex14 from a monomeric soluble form to an oligomeric form entrenched in the peroxisomal membrane. The matrix protein separates from Pex5 and is released into the peroxisomal matrix following the formation of the matrix protein-Pex5-Pex14 complex. The membrane proteins Pex2, Pex10, and Pex1 then ubiquitinylate Pex5, which causes it to be returned to the cytoplasm.Cells from Pex12 and Pex3 deficient patients were stained with fluorescent antibodies to PMP70 or with fluorescent antibodies to catalase, a peroxisomal matrix protein, then viewed in a fluorescence microscope. What are observations possible?a)In wild-type cells, peroxisomal membrane and matrix proteins are NOT visible as bright foci in numerous peroxisomal bodies.b)In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is localized normally to peroxisomal bodiesc)In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is NOT localized normally to peroxisomal bodiesd)In cells from a Pex3-deficient patient, peroxisomal membranes CANassemble, and as a consequence, peroxisomal bodies do not form. Thus both catalase and PMP70 are mis-localized to the cytosol.Correct answer is option 'B'. Can you explain this answer? theory, EduRev gives you an
ample number of questions to practice Catalase and other PTS1-bearing proteins, such as PMP70, are imported into the peroxisomal matrix commencing in the cytosol. For the correct insertion of proteins like PMP70 into the developing peroxisomal membrane, a complex of Pex3 and Pex16 is necessary. First, PTS1 interacts to the Pex5 receptor that has the extraordinary ability to transform in a combination with the membrane protein Pex14 from a monomeric soluble form to an oligomeric form entrenched in the peroxisomal membrane. The matrix protein separates from Pex5 and is released into the peroxisomal matrix following the formation of the matrix protein-Pex5-Pex14 complex. The membrane proteins Pex2, Pex10, and Pex1 then ubiquitinylate Pex5, which causes it to be returned to the cytoplasm.Cells from Pex12 and Pex3 deficient patients were stained with fluorescent antibodies to PMP70 or with fluorescent antibodies to catalase, a peroxisomal matrix protein, then viewed in a fluorescence microscope. What are observations possible?a)In wild-type cells, peroxisomal membrane and matrix proteins are NOT visible as bright foci in numerous peroxisomal bodies.b)In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is localized normally to peroxisomal bodiesc)In cells from a Pex12-deficient patient, catalase is distributed uniformly throughout the cytosol, whereas PMP70 is NOT localized normally to peroxisomal bodiesd)In cells from a Pex3-deficient patient, peroxisomal membranes CANassemble, and as a consequence, peroxisomal bodies do not form. Thus both catalase and PMP70 are mis-localized to the cytosol.Correct answer is option 'B'. Can you explain this answer? tests, examples and also practice Software Development tests.

|
Explore Courses for Software Development exam
|

|
Signup for Free!
Signup to see your scores go up within 7 days! Learn & Practice with 1000+ FREE Notes, Videos & Tests.
























