Structure and synthesis of nucleic acids & proteins | Botany Optional for UPSC PDF Download
| Table of contents |

|
| Nucleic Acid Metabolism |

|
| Replication |

|
| RNA Metabolism |

|
| Protein Synthesis |

|
| Protein Synthesis Definition |

|
| Prokaryotic vs. Eukaryotic Protein Synthesis |

|
| Protein Biosynthesis Steps |

|
Nucleic Acid Metabolism
DNA Metabolism
In eukaryotic cells, DNA metabolism encompasses three primary processes: replication, repair, and recombination. These processes are managed by specialized cellular machinery and are essential for maintaining the integrity of genetic information. Accurate DNA replication is crucial to ensure the faithful transmission of genetic material, while mechanisms are in place to rectify errors occurring during replication or repair DNA damage that may arise post-replication. Recombination between genomes is another vital mechanism, facilitating genetic diversity within a species and aiding DNA repair.
Replication
Basic Mechanisms
- DNA replication is a semi-conservative process where the original DNA molecule's two strands are separated, and new complementary strands are independently created, resulting in two identical copies of the initial DNA. Each copy consists of one parental strand and one newly synthesized strand. Replication commences at a specific location on the chromosome known as the origin and proceeds bidirectionally along the strand, concluding precisely at a defined endpoint. In the case of circular chromosomes, the termination point is reached when the two elongating chains meet, and specific proteins facilitate their connection. DNA polymerases are incapable of initiating replication at the DNA strand's end; they can only extend preexisting short RNA primers, referred to as oligonucleotide fragments. Therefore, in linear chromosomes, specialized mechanisms are in place to initiate and terminate DNA synthesis, preventing information loss.
- The replication of the two DNA strands follows different processes influenced by the direction of the phosphodiester bond. The leading strand is continuously replicated by adding individual nucleotides to its 3' end. In contrast, the lagging strand is synthesized in a disjointed manner: short RNA primers are initially laid down, and then DNA polymerase fills in the gaps, consistently adding bases in the 5' to 3' direction. The brief RNA fragments produced during lagging strand replication are degraded when no longer required. An enzyme called DNA ligase joins the two newly synthesized DNA segments. This enables replication to proceed bidirectionally, resulting in two leading and two lagging strands extending outward from the origin.
Enzymes of Replication
- Replication involves several enzymes and proteins that collectively ensure the accurate duplication of DNA. DNA polymerase is a critical enzyme that adds individual nucleotides to the 3' end of an RNA or DNA molecule during replication. In the prokaryote E. coli, three DNA polymerases exist: one is responsible for chromosome replication, while the other two are involved in repairing damaged DNA. In eukaryotes, this process is even more complex. Human cells, for instance, have more than five different DNA polymerases identified. These various polymerases have specialized roles, including synthesizing the leading and lagging strands during DNA replication and participating in the replication of mitochondrial DNA. Others are primarily engaged in repairing damaged DNA.
- Several additional proteins play essential roles in the replication process. DNA helicases assist in separating the two strands of DNA, while single-stranded DNA binding proteins stabilize these strands during the initial unwinding phase before replication. The unwinding of the DNA double helix introduces strain, leading to supercoiling, which is subsequently alleviated by enzymes known as topoisomerases (as mentioned in the context of supercoiling). Another crucial component is primase, a specialized RNA polymerase that synthesizes the RNA primers necessary to initiate transcription at the replication origin. DNA ligase, on the other hand, seals the nicks created between individual DNA fragments, ensuring the continuity of the newly synthesized DNA strands.
- In eukaryotes, the ends of linear chromosomes contain special sequences called telomeres, which serve as protective caps. Telomeres are generated by the action of a unique DNA polymerase known as telomerase. Telomerase includes an RNA component that serves as a template, guiding the synthesis of the exact telomeric DNA sequence. This repetitive sequence is added multiple times to the ends of chromosomes, counteracting the natural shortening of DNA that occurs during replication and cell division.
- Specialized mechanisms are also employed for the replication of single-stranded viral genomes, mitochondrial genomes, and certain viral genomes. Some viruses, like adenovirus, utilize a nucleotide covalently linked to a protein as a primer for replication. This protein remains attached to the DNA after replication. Other single-stranded viruses employ a rolling circle mechanism for replication. In this process, a double-stranded copy of the viral genome is initially generated, followed by the continuous copying of the nonviral strand. This results in long single-stranded DNA, from which complete viral DNA strands are excised with the assistance of specialized nucleases.
Recombination
- Recombination serves as the primary mechanism for introducing genetic diversity into populations. For instance, during meiosis, the process responsible for producing sex cells like sperm and eggs, homologous chromosomes, which are inherited from both the mother and father, pair up and undergo recombination, a process also known as crossing-over. During recombination, the two DNA molecules are fragmented, and segments of the chromosome that share similarity are exchanged, resulting in the creation of two new chromosomes, each a combination of the original ones. These paired chromosomes eventually separate, ensuring that each sperm or egg receives one of the newly shuffled chromosomes. When fertilization occurs, the standard genetic complement of two copies of each chromosome is reestablished.
- Recombination can be classified into two main types: general and site-specific recombination. General recombination typically involves the breaking and rejoining of DNA strands at regions that are either identical or very similar in sequence. This process is observed in various situations, such as during viral infections, bacterial conjugation (the transfer of genetic material between bacterial cells), DNA transformation (the direct introduction of foreign DNA into cells), and certain DNA repair mechanisms. Site-specific recombination, on the other hand, takes place at specific sites within DNA molecules, often involving the insertion of DNA sequences. Site-specific recombination plays a role in the spread of DNA segments within genomes and is commonly utilized by viruses and specialized DNA elements known as transposons to replicate and disseminate their genetic material.
General recombination
- General recombination, also known as homologous recombination, is a process that involves two DNA molecules with extensive stretches of similar base sequences. Here's how it works:
- Strand Separation: The DNA molecules are initially nicked to produce single strands. These single strands then invade the other DNA duplex, forming a four-stranded DNA structure. This unique structure is known as a Holliday junction, named after Robin Holliday, who proposed this model in 1964.
- Branch Migration: The Holliday junction moves along the DNA duplex by "unzipping" one strand and forming hydrogen bonds on the second strand. This process is called branch migration and leads to the movement of the junction along the DNA.
- Nick Formation: After branch migration, the two DNA duplexes can be nicked again, allowing them to separate.
- Repair by DNA Ligase: Finally, DNA ligase comes into play to repair the nicks, resulting in two DNA duplexes in which the segment between the two nicks has been replaced.
- Several key proteins and enzymes are involved in general recombination:
- RecA: RecA is a crucial enzyme that catalyzes the strand invasion process. It coats single-stranded DNA and facilitates its pairing with a double-stranded DNA molecule containing the same sequence, creating a loop structure.
- RecBC: Another important protein is RecBC, which operates at the free ends of DNA. RecBC catalyzes an unwinding-rewinding reaction as it moves along the DNA molecule. This process generates a loop behind the enzyme, making it easier for subsequent pairing with another DNA molecule.
- Other Proteins: Various other proteins play essential roles in recombination, such as single-stranded DNA binding proteins that stabilize single-stranded DNA, DNA polymerase to repair any gaps that may form, and DNA ligase to reseal the nicks after recombination is complete.
- It's worth noting that during recombination between two DNA regions that are similar but not identical, the initial product may contain mismatched bases at some positions in the helix. This situation is called a "heteroduplex." In meiosis, a round of replication is typically required to properly match the mosaic chromosomes produced by recombination. Cells contain enzymes that can recognize and repair these mismatches. In some cases, the initial products of recombination are repaired before replication, leading to a phenomenon known as gene conversion.
- Recombination can also serve as a mechanism to repair DNA lesions. When one chromosome in a pair is irreversibly damaged, recombination allows for the copying and insertion of genetic information from the intact chromosome to replace the damaged section. This process involves base-pairing between sequences flanking the damage on the damaged chromosome and the corresponding sequences on the intact chromosome, enabling the correct sequence to be replicated and the lesion to be repaired.
Site-specific recombination
- Site-specific recombination is a process in which specific short sequences in DNA are recognized by proteins and can be inserted, removed, or inverted, leading to significant changes in the genome. This mechanism is responsible for reshaping genomes and can be seen in various organisms, including plants and humans.
- One example of site-specific recombination is the integration of DNA from bacteriophage λ into the chromosome of E. coli. This process involves the formation of a circular bacteriophage λ DNA, which is then cleaved at a specific site by an enzyme called λ-integrase. A similar site on the bacterial chromosome is also cut by integrase, and the two ends can be rejoined, resulting in the insertion of the phage DNA into the bacterial chromosome. This integrated phage can remain inactive until signals trigger its release and resumption of its life cycle. The process of excision, in which the phage DNA is removed from the bacterial chromosome, is facilitated by another protein called excisionase.
- Transposons, or "jumping genes," also exhibit a similar process of integration and excision. These elements can move from one location in a genome to another and are always maintained in an integrated site. Transposons encode an enzyme called transposase that cleaves the ends of the transposon and its target site. There are two types of transposition: simple movement to another site in the chromosome and replicative transposition, which involves replication of the transposon before insertion, generating a second copy. Replicative transposition is responsible for the widespread spread of transposable elements in many organisms.
- Transposons can carry additional genes, such as antibiotic resistance factors, which provide a selective advantage to the host organism. Antibiotic resistance genes are often found on transposable elements that have moved into plasmids and can be easily transferred between organisms. The indiscriminate use of antibiotics can promote the accumulation of drug-resistant plasmids and strains.
Repair
- Ensuring the integrity of DNA is of utmost importance for the proper functioning of cells throughout their lifespan and for the accurate transmission of genetic information from one generation to the next. To maintain DNA integrity, repair processes are constantly active, monitoring the DNA for any damage or abnormalities and activating the appropriate repair enzymes. While we discussed recombination mechanisms for repairing severe DNA damage like pyrimidine dimers or gaps in a previous section, there are various other repair mechanisms at play.
- One crucial repair mechanism we'll focus on is mismatch repair, which has been extensively studied in E. coli. This system is guided by the presence of a methyl group within the GATC sequence on the template strand. Similar mismatch repair systems are also present in eukaryotes, although the template strand in this case is not marked by methyl groups. Interestingly, mutations within genes responsible for the human mismatch repair system are known to contribute to many cancer types. When the mismatch repair system is compromised, mutations accumulate rapidly and eventually impact the genes responsible for cell division control. Consequently, cells begin to divide uncontrollably, leading to the development of cancer.
- After DNA replication is complete, the most common type of damage to nucleic acids involves changes in the normal A, C, G, and T bases, resulting in chemically modified bases that differ significantly from their natural counterparts. The exceptions are the conversion of cytosine to uracil and 5-methylcytosine to thymine, which create G:U or G:T mismatches. Specific enzymes known as DNA glycosylases can identify uracil or thymine in DNA and selectively remove these bases by breaking the bond between the base and the deoxyribose sugar. Many of these enzymes are designed to target the various chemically modified bases that may be present in DNA.
- Another common method for repairing DNA damage is through an excision repair pathway. Enzymes are capable of recognizing damaged sections within DNA, often by detecting alterations in the DNA's structure. Subsequently, these enzymes make incisions on either side of the damaged site, allowing for the removal of a small, single-stranded DNA segment. DNA polymerase and DNA ligase then collaborate to repair the single-stranded gap. In all these repair systems, the presence of an abnormal base indicates which strand needs repair, and the complementary strand serves as the template to ensure the accuracy of the repair process.
RNA Metabolism
RNA metabolism is a vital process that bridges the gap between the genetic information stored in DNA and the functional activities of cells. Various types of RNA molecules, such as rRNAs and snRNAs, become part of complex ribonucleoprotein structures with specialized roles in the cell. Others, like tRNAs and mRNAs, play critical roles in protein synthesis, with mRNA directing protein production by ribosomes. RNA metabolism unfolds in three distinct phases: transcription, processing of precursor RNAs into functional forms, and RNA degradation for recycling of bases. Transcription entails copying specific DNA segments (genes) into RNA molecules, which encode proteins or fulfill structural/catalytic roles. Translation involves the decoding of mRNA information on ribosomes to build proteins. Notably, there are distinctions in both transcription and translation between prokaryotic and eukaryotic organisms.
Transcription
- In transcription, small segments of DNA are transcribed into RNA by the enzyme RNA polymerase in a tightly regulated process. This process begins by identifying a specific DNA sequence known as a promoter, signaling the gene's start. At this point, the DNA strands separate, and RNA polymerase starts copying from a specific spot on one DNA strand using a ribonucleoside 5′-triphosphate to initiate the RNA chain. Ribonucleoside triphosphates are used as substrates, and they are incorporated into the growing RNA chain by breaking their high-energy phosphate bond.
- The complementary base pairing rules of DNA dictate the sequence of ribonucleotides: C in DNA leads to G in RNA, G to C, T to A, and A to U. The synthesis continues until a termination signal is encountered, prompting RNA polymerase to detach from DNA, releasing the RNA molecule. Sometimes, this RNA is the final mRNA, while in other cases, it is a pre-mRNA requiring further processing before translation. In prokaryotes, "operators" upstream of genes can bind repressor proteins, blocking RNA polymerase's access and preventing transcription. Activator proteins can also promote transcription by binding to specific signals ahead of certain prokaryotic genes.
- Transcription is more intricate in eukaryotes. Eukaryotic RNA polymerase is a more complex enzyme, and many accessory factors, called transcription factors, regulate promoter efficiency, responding to cellular signals indicating the need for transcription. Numerous transcription factors may be required for efficient transcription in human genes. Transcription factors can activate or repress gene expression in eukaryotes.
- During transcription, only one DNA strand is usually copied—the template strand. The resulting RNA molecules are single-stranded. The DNA strand corresponding to the mRNA is called the coding or sense strand, and it can vary from one gene to another. In eukaryotes, the initial product of transcription is pre-mRNA, which undergoes extensive splicing before maturing into mRNA ready for ribosome-mediated translation.
Translation
- Translation utilizes the information encoded in mRNA's nucleotide sequence to guide protein synthesis. This process occurs on ribosomes, complex cellular structures containing RNA and proteins. In prokaryotes, ribosomes bind to mRNA while transcription is ongoing. Near the mRNA's 5′ end, a short sequence called the ribosome binding site (Shine-Dalgarno sequence) marks the translation's starting point. This sequence is located typically five to eight bases upstream of the initiation codon. The mRNA sequence is read in triplets (codons) from the 5′ to 3′ end, with each amino acid added to the growing chain by its respective aminoacyl tRNA. Translation ceases when a termination codon (UAG, UAA, or UGA) is encountered. Special release factors associate with the ribosome in response to these codons, leading to the dissociation of the newly synthesized protein, tRNAs, and mRNA. The ribosome is then free to engage with another mRNA molecule.
- In eukaryotes, protein synthesis follows the same principles, but ribosomes are more complex. Like in prokaryotes, translation initiates through the interaction of the signal sequence with the 3′ end of the small subunit rRNA during the formation of the initiation complex.
- Accuracy during protein synthesis, while important, is not as critical as during replication. Occasionally, mistranslated proteins do not fold correctly and are degraded. However, ribosomes have proofreading mechanisms to ensure accurate pairing between mRNA codons and tRNA anticodons.
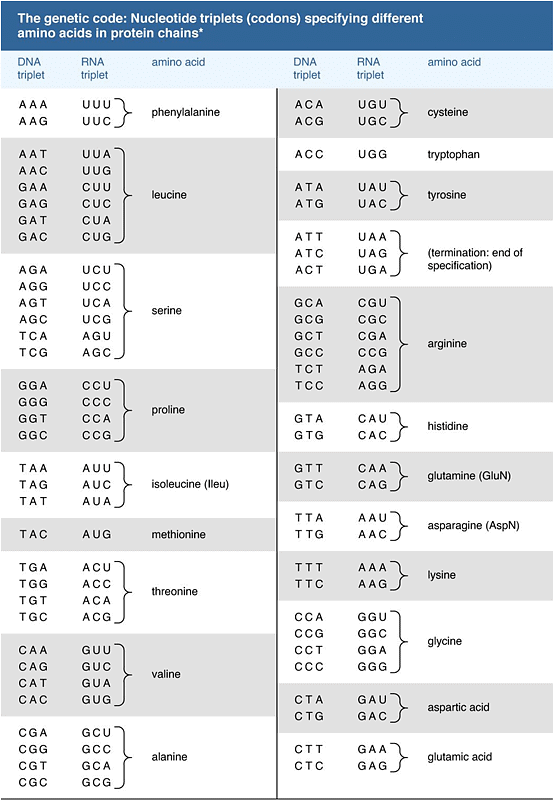
- A significant milestone in molecular biology was the discovery of the genetic code, achieved during the 1960s by scientists like Har G. Khorana and Marshall W. Nirenberg, who shared a Nobel Prize in 1968. This code dictates that the 20 amino acids are encoded by 61 codons, allowing some degeneracy in coding. The genetic code appears nearly universal among various organisms, with exceptions found in mitochondrial DNA and certain bacteria. The structure of the genetic code suggests it may have evolved from a more primitive code involving 16 dinucleotides, though the specifics remain speculative.
Protein Synthesis
Protein synthesis, a fundamental biological process, plays a pivotal role in building the diverse array of proteins essential for the functioning of living organisms. This article delves into the intricacies of protein synthesis, shedding light on its definition, etymology, and the contrasting processes in prokaryotes and eukaryotes. It also explores the genetic code, the key players in protein synthesis (mRNA, tRNA, and rRNA), and the critical steps involved in protein biosynthesis.
Protein Synthesis Definition
- Protein synthesis, often abbreviated as protein biosynthesis, refers to the intricate mechanism by which proteins are created within the biological systems. This process encompasses various biochemical events, including amino acid synthesis, transcription, translation, and post-translational events.
- Amino Acid Synthesis: One of the initial steps in protein synthesis involves the production of amino acids from carbon sources like glucose. While some amino acids are synthesized within the body, others are acquired through dietary sources.
- Transcription and Translation: Inside the cells, proteins are generated through transcription and translation processes. Transcription is the process through which the mRNA template is transcribed from DNA, serving as a blueprint for the subsequent step - translation. During translation, amino acids are sequentially linked together based on the genetic code.
- Post-Translational Events: After translation, the newly formed protein undergoes further processing, including proteolysis, post-translational modification, and protein folding. These events are vital for ensuring proper protein function.
Etymology
The term "protein" has its origins in Late Greek, specifically prōteios and prōtos, which signify "first." "Synthesis" is derived from the Greek word sunthesis, originating from suntithenai, meaning "to put together." An alternative term for protein synthesis is protein biosynthesis.
Prokaryotic vs. Eukaryotic Protein Synthesis
Proteins are crucial biomolecules required by all living organisms, serving various functions such as structural support and catalyzing biochemical reactions.
However, protein synthesis in prokaryotes and eukaryotes differs significantly:
- Prokaryotic Protein Synthesis: In prokaryotes, such as bacteria, protein synthesis occurs in the cytoplasm. Notably, translation begins even before transcription of mRNA is complete.
- Eukaryotic Protein Synthesis: In eukaryotes, protein synthesis initiates in the nucleus, where a transcript (mRNA) of the DNA coding region is created. This mRNA then exits the nucleus to reach the ribosomes, where translation into a protein with a specific amino acid sequence takes place.
Here is a table highlighting the distinctions between prokaryotic and eukaryotic protein synthesis: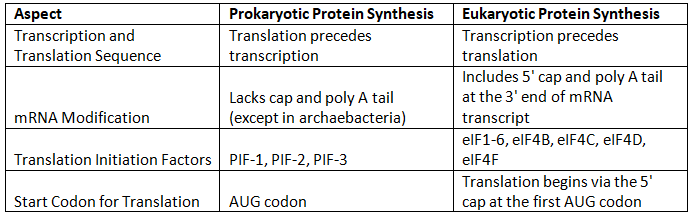
Genetic Code
The genetic code is a pivotal system that orchestrates the intricate components of protein synthesis, encompassing DNA, mRNA, and tRNA. Codons, comprising trinucleotides, specify particular amino acids. For example, the codon GCC codes for alanine, GUU for valine, and UAA serves as a stop codon. The mRNA codon complements the trinucleotide, known as an anticodon, present in tRNA.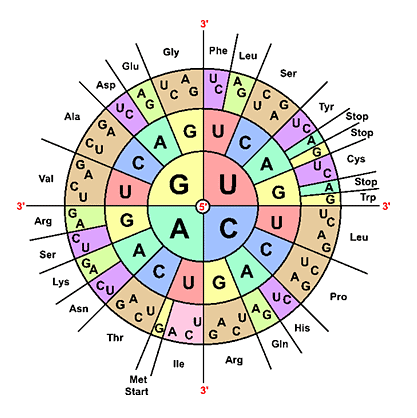
mRNA, tRNA, and rRNA
Three major types of RNA play central roles in protein synthesis:
- mRNA (Messenger RNA): mRNA carries the genetic code for protein synthesis. It consists of various regions, including the 5' cap, 5' untranslated region (UTR), coding region, 3' UTR, and poly(A) tail. The coding region holds the DNA segment for gene expression, commencing with a start codon and concluding with a stop codon.
- tRNA (Transfer RNA): tRNA transfers specific amino acids to the ribosome, where they are incorporated into the growing amino acid chain. It comprises an anticodon arm and an acceptor stem, where amino acids are attached.
- rRNA (Ribosomal RNA): Unlike mRNA and tRNA, rRNA does not carry genetic information but serves as an integral part of ribosomes. Ribosomes are cellular structures where protein synthesis occurs. Prokaryotes have 70S ribosomes, while eukaryotes possess 80S ribosomes, each composed of two subunits with different sizes. The larger subunit acts as a ribozyme, catalyzing peptide bond formation between amino acids. rRNA features three binding sites: A (aminoacyl), P (peptidyl), and E (exit).
Protein Biosynthesis Steps
Protein biosynthesis involves several major steps, including transcription, translation, and post-translation modifications. Let's focus on the transcription step in detail.
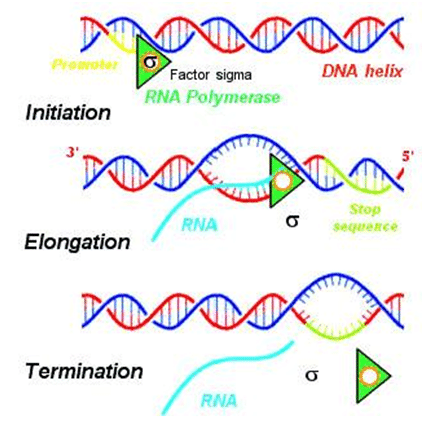
Transcription
Transcription is the initial stage of protein biosynthesis where an mRNA molecule is synthesized based on the information encoded in DNA. This process provides a template for subsequent translation and is carried out by an enzyme called RNA polymerase.
In transcription, the following key steps occur:
- Step 1: Initiation: Initiation is the first phase of transcription. During this step, RNA polymerase, aided by specific transcription factors, binds to the DNA's promoter region. This binding leads to the unwinding of the DNA, creating a transcription bubble. Within this bubble, a transcription start site on the DNA binds to the RNA polymerase, along with the initiating and extending ribonucleoside triphosphates (NTPs). Initially, there are some abortive cycles of synthesis, resulting in the production of short mRNA transcripts, typically containing 2 to 15 nucleotides.
- Step 2: Promoter Escape: After initiation, the RNA polymerase needs to escape from the promoter region to proceed to the next phase, elongation.
- Step 3: Elongation: During elongation, the RNA polymerase moves along the DNA template strand, base-pairing with complementary nucleotides on the noncoding DNA strand. This process results in the formation of an mRNA transcript that carries a copy of the coding strand of DNA. Notably, uracil (U) is incorporated instead of thymine (T) in the mRNA. The sugar-phosphate backbone of the mRNA forms as RNA polymerase progresses.
- Step 4: Termination: Termination is the final phase of transcription. In this stage, the hydrogen bonds holding together the RNA-DNA helix break, and the newly synthesized mRNA molecule is released. In eukaryotes, the mRNA transcript undergoes additional processing steps, including polyadenylation (adding a poly-A tail), capping, and splicing to ensure its functionality.
Translation
Translation is a critical step in protein biosynthesis where amino acids are linked together in a specific order according to the instructions encoded in mRNA.
This process occurs in the cytoplasm, where ribosomes are located, and it involves four phases:
- Activation: During activation, an amino acid is covalently bonded to its corresponding transfer RNA (tRNA), forming aminoacyl-tRNA. This step ensures that each amino acid is linked to the appropriate tRNA molecule, which will later recognize the mRNA codon specifying that amino acid.
- Initiation: In the initiation phase, the small subunit of the ribosome binds to the 5′ end of the mRNA molecule. This binding is facilitated by initiation factors. This step ensures that translation begins at the correct starting point on the mRNA.
- Elongation: Elongation is the phase where the next aminoacyl-tRNA molecule in line binds to the ribosome. This binding occurs with the help of GTP (guanosine triphosphate) and an elongation factor. The ribosome moves along the mRNA, matching each codon with the appropriate aminoacyl-tRNA and linking the amino acids together to form a growing polypeptide chain.
- Termination: In the termination phase, the ribosome reaches a stop codon on the mRNA, signaling the end of translation. When a stop codon enters the ribosome's A site, it does not correspond to any aminoacyl-tRNA. Instead, it is recognized by release factors, leading to the release of the completed polypeptide chain from the ribosome.
Post-translation Events
Following protein synthesis, several post-translation events occur to ensure the proper structure and function of the newly synthesized protein:
- Proteolysis: Proteolysis involves the cleavage of proteins by enzymes called proteases. This process can remove specific amino acid residues from the N-terminal, C-terminal, or internal regions of the polypeptide. Proteolysis plays a role in activating certain proteins and regulating their function.
- Post-translational Modification: Post-translational modification refers to enzymatic changes made to a polypeptide chain after translation and peptide bond formation. These modifications can include the addition of chemical groups (e.g., phosphorylation, acetylation, glycosylation) to amino acid side chains or the cleavage of specific peptide bonds. These modifications can affect protein stability, localization, and activity.
- Protein Folding: Protein folding is the process by which the polypeptide chain adopts its secondary and tertiary structures, ultimately forming the functional, three-dimensional protein. Proper protein folding is essential for the protein to carry out its biological functions correctly. Molecular chaperones often assist in the folding process to prevent misfolding and aggregation.
|
179 videos|143 docs
|














