More Complex Spin-Spin Splitting Patterns | Chemistry Optional Notes for UPSC PDF Download
More Complex Spin-Spin Splitting Patterns
In the 1H NMR spectra we’ve seen so far, the chemical shifts of different protons have been distinct and the spin–spin splitting patterns have been straightforward. It often happens, however, that different kinds of hydrogens in a molecule have accidentally overlapping signals. The spectrum of toluene (methylbenzene) in Figure 13.13, for example, shows that the five aromatic ring protons give a complex, overlapping pattern, even though they aren’t all equivalent.
Yet another complication in 1H NMR spectroscopy arises when a signal is split by two or more nonequivalent kinds of protons, as is the case with trans-cinnamaldehyde, isolated from oil of cinnamon (Figure 13.14). Although the n + 1 rule predicts splitting caused by equivalent protons, splittings caused by nonequivalent protons are more complex.
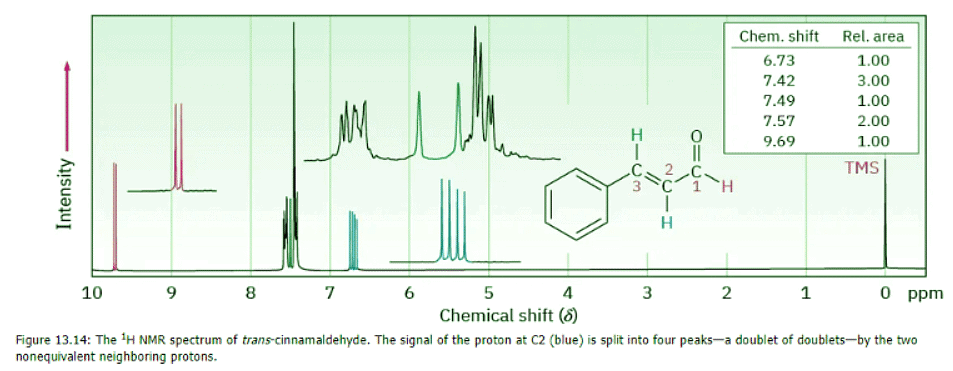
To understand the 1H NMR spectrum of trans-cinnamaldehyde, we have to isolate the different parts and look at the signal of each proton individually.
- The five aromatic proton signals (black in Figure 13.14) overlap into a complex pattern with a large peak at 7.42 δ and a broad absorption at 7.57 δ.
- The aldehyde proton signal at C1 (red) appears in the normal downfield position at 9.69 δ and is split into a doublet with J = 6 Hz by the adjacent proton at C2.
- The vinylic proton at C3 (green) is next to the aromatic ring and is therefore shifted downfield from the normal vinylic region. This C3 proton signal appears as a doublet centered at 7.49 δ. Because it has one neighbor proton at C2, its signal is split into a doublet, with J = 12 Hz.
- The C2 vinylic proton signal (blue) appears at 6.73 δ and shows an interesting, four-line absorption pattern. It is coupled to the two nonequivalent protons at C1 and C3 with two different coupling constants: J1-2 = 6 Hz and J2-3 = 12 Hz.
A good way to understand the effect of multiple coupling, such as that occurring for the C2 proton of trans-cinnamaldehyde, is to draw a tree diagram, like that in Figure 13.15. The diagram shows the individual effect of each coupling constant on the overall pattern. Coupling with the C3 proton splits the signal of the C2 proton in trans-cinnamaldehyde into a doublet with J =12 Hz. Further coupling with the aldehyde proton then splits each peak of the doublet into new doublets with J = 6 Hz, and we therefore observe a four-line spectrum for the C2 proton.
One further trait evident in the cinnamaldehyde spectrum is that the four peaks of the C2 proton signal are not all the same size. The two left-hand peaks are somewhat larger than the two right-hand peaks. Such a size difference occurs whenever coupled nuclei have similar chemical shifts—in this case, 7.49 δ for the C3 proton and 6.73 δ for the C2 proton. The peaks nearer the signal of the coupled partner are always larger, and the peaks farther from the signal of the coupled partner are always smaller. Thus, the left-hand peaks of the C2 proton multiplet at 6.73 δ are closer to the C3 proton absorption at 7.49 δ and are larger than the right-hand peaks. At the same time, the right-hand peak of the C3 proton doublet at 7.49 δ is larger than the left-hand peak because it is closer to the C2 proton multiplet at 6.73 δ. This skewing effect on multiplets can often be useful because it tells where to look in the spectrum to find the coupled partner: look in the direction of the larger peaks.
FAQs on More Complex Spin-Spin Splitting Patterns - Chemistry Optional Notes for UPSC
| 1. What are spin-spin splitting patterns in nuclear magnetic resonance (NMR) spectroscopy? |  |
| 2. How do spin-spin splitting patterns provide information about molecular structure? |  |
| 3. What factors influence the complexity of spin-spin splitting patterns? |  |
| 4. How can one interpret spin-spin splitting patterns in NMR spectra? |  |
| 5. Can spin-spin splitting patterns be used to distinguish between different isomers or conformers of a molecule? |  |

















