Transcription, Genetic Code & Translation | Biology Class 12 - NEET PDF Download
| Table of contents |

|
| Transcription |

|
| Genetic Code |

|
| Mutations and Genetic Code |

|
| tRNA: The Adapter Molecule |

|
| Translation |

|
Transcription
Transcription is the process of copying genetic information from DNA into RNA. It follows the principle of complementarity, where adenosine pairs with uracil instead of thymine. Unlike replication, which duplicates the entire DNA, transcription only copies a specific segment of DNA from one strand into RNA.
Transcription Unit
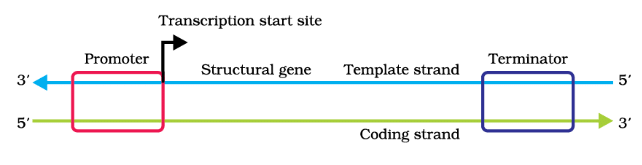 Transcription Unit
Transcription Unit
- Definition: A transcription unit in DNA consists of three main regions: a promoter, a structural gene, and a terminator.
- Strands: The two strands of DNA in a transcription unit have opposite polarities. The strand with a 3'→5' polarity acts as the template strand, while the 5'→3' strand, which has the same sequence as the RNA (except for thymine), is called the coding strand.
- Example: In a hypothetical transcription unit,
template strand : 3' -ATGCATGCATGCATGCATGCATGC-5'
coding strand: 5' -TACGTACGTACGTACGTACGTACG-3'.
The RNA transcribed from this DNA would have the same sequence as the coding strand, with uracil replacing thymine. - Promoter and Terminator: The promoter is located upstream (5' end) of the structural gene and provides a binding site for RNA polymerase. The terminator is located downstream (3' end) and marks the end of transcription. The presence of the promoter determines the template and coding strands.
- Coding Strand: The coding strand, despite not coding for anything, serves as a reference point for defining the transcription unit. If the coding and template strands were reversed, the roles of RNA polymerase and the transcription process would change.
- Regulatory Sequences: Additional regulatory sequences may be present further upstream or downstream of the promoter and can influence gene expression.
Transcription Unit and the Gene
Definition of a Gene
- A gene is considered the fundamental unit of inheritance. While it is clear that genes are located on DNA, precisely defining a gene based on DNA sequence can be challenging.
- Genes can also include DNA sequences that code for non-protein molecules, such as tRNA or rRNA.
Cistron and Structural Genes
- A cistron refers to a segment of DNA that codes for a specific polypeptide.
- Based on this definition, structural genes within a transcription unit can be classified as monocistronic or polycistronic.
- Monocistronic genes, common in eukaryotes, contain a single coding sequence.
- Poly cistronic genes, typical in bacteria or prokaryotes, can code for multiple polypeptides.
Monocistronic Structural Genes in Eukaryotes
- In eukaryotes, monocistronic structural genes often have interrupted coding sequences, meaning the genes are "split."
- The coding sequences, or expressed sequences, are known as exons.
- Exons are the sequences that appear in the mature or processed RNA.
- Exons are separated by introns.
- Introns, or intervening sequences, do not appear in the mature or processed RNA.
Split-Gene Arrangement and Regulatory Sequences
- The presence of introns and the split-gene arrangement make it difficult to define a gene solely based on a DNA segment.
- The inheritance of a trait is also influenced by promoter and regulatory sequences associated with a structural gene.
- Sometimes, these regulatory sequences are referred to as regulatory genes, even though they do not code for any RNA or protein.
Types of RNA and the Process of Transcription
Transcription is the process by which RNA is synthesized from a DNA template. In bacteria, there are three major types of RNA involved in this process: messenger RNA (mRNA), transfer RNA (tRNA), and ribosomal RNA (rRNA).
Types of RNA in Bacteria
- mRNA (messenger RNA): Serves as the template for protein synthesis, carrying the genetic information from DNA to the ribosomes.
- tRNA (transfer RNA): Brings amino acids to the ribosomes and helps decode the mRNA sequence into a polypeptide chain.
- rRNA (ribosomal RNA): Forms the structural and catalytic components of ribosomes, playing a crucial role in protein synthesis.
1. Transcription Process in Bacteria
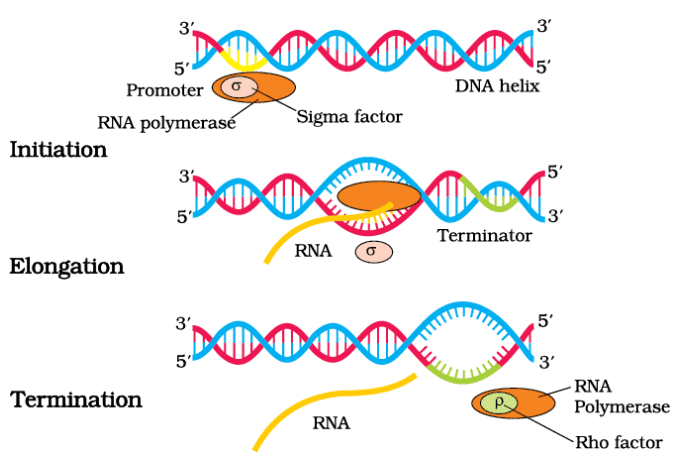 Process of Transcription in Bacteria
Process of Transcription in Bacteria
- Initiation: RNA polymerase binds to the promoter region of the DNA and unwinds the DNA double helix.
- Elongation: RNA polymerase synthesizes the RNA strand by adding nucleotides complementary to the DNA template strand.
- Termination: RNA polymerase reaches the terminator region, causing the newly synthesized RNA to detach from the enzyme and the DNA.
Role of RNA Polymerase
- RNA polymerase is responsible for catalyzing the elongation phase of transcription. It requires the assistance of initiation factors (σ) for starting transcription and termination factors (ρ) for ending the process.
- In bacteria, mRNA does not undergo processing and can be translated immediately after transcription. Transcription and translation occur simultaneously in the same cellular compartment.
2. Transcription in Eukaryotes
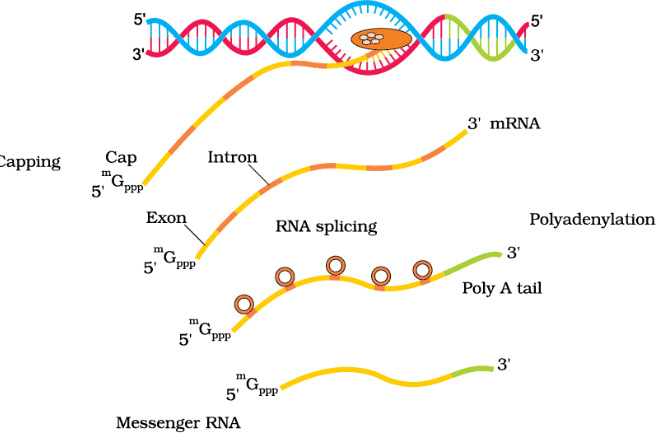 Process of Transcription in Eukaryotes
Process of Transcription in Eukaryotes
In eukaryotic cells, transcription is more complex due to the presence of three different RNA polymerases, each with specific functions:
- RNA Polymerase I: Transcribes rRNA (28S, 18S, and 5.8S).
- RNA Polymerase II: Transcribes precursor mRNA (hnRNA).
- RNA Polymerase III: Transcribes tRNA, 5S rRNA, and small nuclear RNAs (snRNAs).
Processing of hnRNA
- hnRNA undergoes splicing to remove introns and join exons in a specific order.
- Capping involves adding a methyl guanosine triphosphate cap to the 5'-end of hnRNA.
- Tailing involves adding 200-300 adenylate residues to the 3'-end of hnRNA.
- After processing, hnRNA becomes mature mRNA, which is transported out of the nucleus for translation.
Significance of RNA Processing
- The presence of introns and the process of splicing are believed to be ancient features of the genome, reflecting the dominance of RNA-based processes in early life forms.
- Understanding RNA and RNA-dependent processes is crucial for comprehending the complexities of gene expression and regulation in living organisms.
Genetic Code
Introduction
During the processes of replication and transcription, a nucleic acid is copied to produce another nucleic acid. These processes are relatively straightforward to understand based on the principle of complementarity. However, translation involves the transfer of genetic information from a polymer of nucleotides to synthesize a polymer of amino acids, where no complementarity exists between nucleotides and amino acids.
Despite this lack of theoretical complementarity, there is strong evidence that changes in nucleic acids (genetic material) are responsible for changes in the amino acids of proteins. This led to the development of the genetic code, which directs the sequence of amino acids during protein synthesis.
Determining the biochemical nature of genetic material and the structure of DNA was an exciting achievement, but the proposition and deciphering of the genetic code posed a significant challenge. It required the collaboration of scientists from various disciplines, including physics, organic chemistry, biochemistry, and genetics.
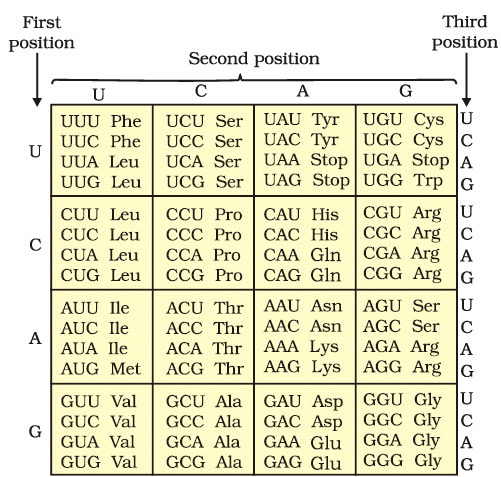 The codons for the various amino acids
The codons for the various amino acids
Triplet Codon Hypothesis
George Gamow, a physicist, proposed the idea that since there are only four bases in nucleic acids and they must code for 20 amino acids, the genetic code should consist of combinations of bases. He suggested that the code is made up of three nucleotides (triplet codons) to account for all 20 amino acids.
This was a bold proposition because a combination of three nucleotides (4 x 4 x 4) would generate 64 codons, exceeding the number needed. Proving that the codon is indeed a triplet was a challenging task.
Contributions of Scientists
Har Gobind Khorana developed chemical methods to synthesize RNA molecules with defined combinations of bases (homopolymers and copolymers), which was crucial for understanding the genetic code. Marshall Nirenberg's cell-free system for protein synthesis played a pivotal role in deciphering the code.
Severo Ochoa's enzyme, polynucleotide phosphorylase, also contributed by polymerizing RNA with defined sequences in a template-independent manner (enzymatic synthesis of RNA). These efforts culminated in the preparation of a checkerboard for the genetic code.
Salient Features of Genetic Code
- Codon Triplet: The genetic code is based on triplets of nucleotides called codons. There are 61 codons that specify amino acids, while 3 codons serve as stop signals, indicating the end of protein synthesis.
- Degeneracy: Some amino acids are encoded by more than one codon, making the code degenerate. For example, multiple codons can specify the same amino acid.
- Contiguous Reading: The codons in mRNA are read in a continuous manner without any punctuation or gaps between them.
- Universality: The genetic code is nearly universal across different organisms. For instance, the codon UUU codes for the amino acid Phenylalanine (Phe) in both bacteria and humans. However, there are exceptions in mitochondrial codons and certain protozoans.
- AUG Codon: The codon AUG serves a dual purpose. It codes for the amino acid Methionine (Met) and also acts as the initiator codon, signaling the start of protein synthesis.
- Stop Codons: The codons UAA, UAG, and UGA function as stop codons, signaling the termination of protein synthesis.
Practice Question
Predicting Amino Acid Sequence from mRNA
Given mRNA Sequence:-AUG UUU UUC UUC UUU UUU UUC
Predict the sequence of amino acids encoded by this mRNA using the genetic code checkerboard.
Predicting mRNA Sequence from Amino Acids
Given Amino Acid Sequence: Met-Phe-Phe-Phe-Phe-Phe-Phe
Predict the corresponding nucleotide sequence in the mRNA. Did you encounter any difficulties in making this prediction? Can you now relate this task to the properties of the genetic code you have learned?
Mutations and Genetic Code
Understanding the relationship between genes and DNA is crucial, and mutation studies play a key role in this. Mutations can have significant effects on DNA, leading to the loss or gain of genes and their associated functions.
One classic example of a mutation is a point mutation, where a single base pair change in the gene for the beta globin chain can alter the amino acid from glutamate to valine, causing sickle cell anemia.
Effects of Point Mutations
- Point mutations can have significant effects on genetic information.
- A classic example is the mutation in the beta globin gene, where a single base pair change alters glutamate to valine, leading to sickle cell anemia.
Frameshift Mutations
- Frameshift mutations occur due to the insertion or deletion of nucleotide bases.
- These mutations can disrupt the reading frame of the genetic code, altering the resulting protein.
Examples of Frameshift Mutations
- Insertion of One or Two Bases: Alters the reading frame from the point of insertion, leading to a different sequence of amino acids.
- Insertion of Three or More Bases: Can insert or delete one or multiple codons without changing the reading frame from that point onward.
tRNA: The Adapter Molecule
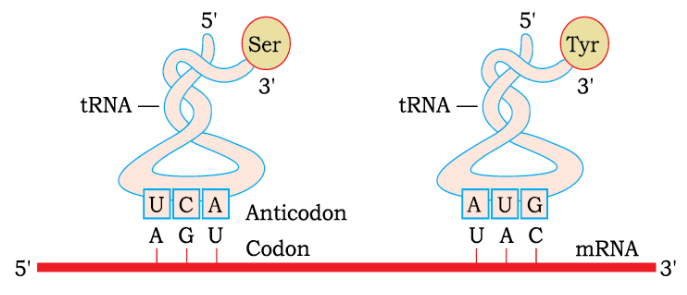
Francis Crick recognized the need for a mechanism to read the genetic code and link it to amino acids, as amino acids alone could not uniquely interpret the code. He proposed the existence of an adapter molecule that would read the code on one end and bind specific amino acids on the other.
tRNA Discovery and Role
- tRNA, initially known as sRNA (soluble RNA), was discovered before the genetic code was proposed.
- However, its role as an adapter molecule was understood much later.
Structure of tRNA
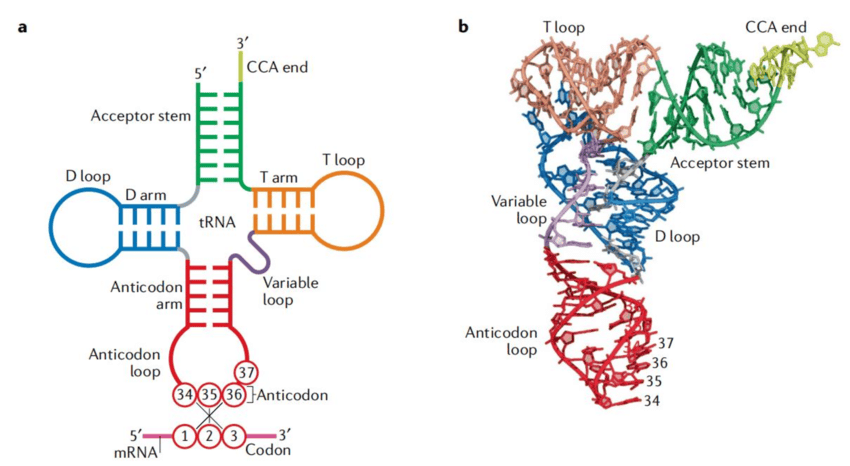 Structure of tRNA
Structure of tRNA
Anticodon Loop: The tRNA has an anticodon loop with bases that are complementary to the genetic code.
Amino Acid Acceptor End: Each tRNA has an amino acid acceptor end where specific amino acids bind.
Specificity: tRNAs are specific to each amino acid, ensuring the correct amino acid is added during protein synthesis.
Initiator tRNA: There is a specific tRNA called initiator tRNA that is involved in the initiation of protein synthesis.
Stop Codons: There are no tRNAs for stop codons, which signal the termination of protein synthesis.
Cloverleaf Structure: The secondary structure of tRNA is often depicted as a cloverleaf.
Actual Structure: In reality, tRNA is a compact molecule that resembles an inverted L shape.
Translation
Translation is the process of converting the genetic information stored in mRNA into a specific sequence of amino acids, ultimately forming a polypeptide chain, which folds into a functional protein.
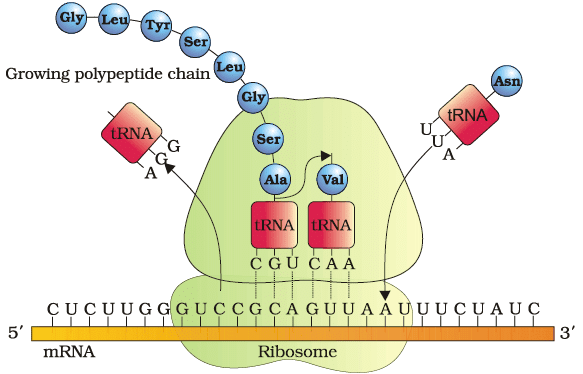 Translation
Translation
Process of Translation
- Initiation: The small subunit of the ribosome binds to the mRNA at the start codon (AUG), which is recognized by the initiator tRNA carrying the amino acid methionine.
- Elongation: Aminoacyl-tRNA complexes, each carrying a specific amino acid, sequentially bind to the codons on the mRNA through complementary base pairing between the tRNA anticodon and the mRNA codon. The ribosome facilitates the formation of peptide bonds between adjacent amino acids, linking them together to form a growing polypeptide chain.
- Termination: When the ribosome encounters a stop codon on the mRNA, a release factor binds to the stop codon, prompting the release of the completed polypeptide chain from the ribosome.
|
59 videos|290 docs|168 tests
|
FAQs on Transcription, Genetic Code & Translation - Biology Class 12 - NEET
| 1. What is the role of RNA polymerase in transcription? |  |
| 2. What are the main stages of transcription? |  |
| 3. How does transcription termination occur? |  |
| 4. What is the function of ribosomal RNA (rRNA) in translation? |  |
| 5. What is the significance of the T-arm in transfer RNA (tRNA)? |  |















