Principles of Biotechnology: Genetic Engineering | Biology Class 12 - NEET PDF Download
| Table of contents |

|
| Genetic Engineering |

|
| Tools of Biotechnology |

|
| 2) Cloning Vectors |

|
| 3) Competent Host |

|
Genetic Engineering
Introduction
Genetic engineering also referred as ‘recombinant DNA technology ’ or ‘gene splicing ’ is one kind of biotechnology involving manipulation of DNA.
Products of Genetic Engineering
It deals with the isolation of useful genes from a variety of sources and the formation of new combinations of DNA (recombinant DNA) for repair, improvement, perfection and matching of a genotype. Thus, genetic engineering may be defined ‘as a technique for artificial and deliberately modifying DNA (gene) to suit human needs’. In genetic engineering breakage of DNA molecule at two desired places is done with the help of restriction endonuclease to isolate a specific DNA segment and then insert it in another DNA molecule at a desired position.
The new DNA molecule is recombinant DNA and the technique is called genetic engineering. Genetic engineering aims at adding, removing or repairing of a part of genetic material. Genetic engineering can be used to improve the quality of human life.
Paul Berg (Father of Genetic Engineering)
He transferred gene of SV-40 virus (simian virus) into E.coli with the help of l – phage (Nobel Prize-1980).
 Paul Berg
Paul Berg
The concept of genetic engineering was the outcome of two very significant discoveries made in bacterial research. These were:
(i) presence of extra chromosomal DNA fragments called plasmids in the bacterial cell,which replicate along with chromosomal DNA of the bacterium.
(ii) presence of enzymes restriction endonucleases which cut DNA at specific sites.
These enzymes are, therefore, called ‘molecular scissors’.
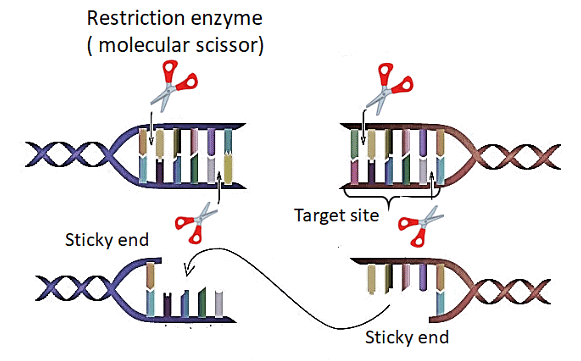
Tools of Biotechnology
Genetic engineering or recombinant DNA technology is possible only if we have some important tools. The following are some important tools of biotechnology:
1) Restriction Enzymes
Restriction enzymes, also known as ‘restriction endonucleases’ are molecular scissors that can cut DNA at specific locations. These are part of a larger class of enzymes called ‘Nucleases’. Nucleases are of two kinds:
- Exonucleases – Enzymes that remove nucleotides from the ends of DNA.
- Endonucleases – Enzymes that make cuts at specific positions in the DNA.
Hind II– the first restriction endonuclease to be isolated, identifies a specific six base pair sequence and always cuts the DNA at a particular point. This specific sequence is the ‘Recognition sequence‘. Today, there are more than 900 restriction enzymes, isolated from about 230 bacterial strains. Each of these enzymes recognizes different recognition sequences. There is a naming convention to name restriction enzymes. Let’s learn this using EcoRI as an example:
- Eco – indicates that this enzyme was isolated from Escherichia coli.
- R – refers to the name of the strain.
- I – is the Roman numeral that indicates the order of isolation of this restriction enzyme from the strain of bacteria.
Mode of Action of Restriction Enzymes
Endonucleases perform their function in the following manner:
- Inspect the length of DNA for the specific recognition sequence.
- Bind the DNA at the recognition sequence.
- Cut the two strands of DNA at specific points in their sugar-phosphate backbone.
Restriction enzymes recognize specific palindromic nucleotide sequences in the DNA. A palindrome is a group of letters that produce the same word when reading forward or backwards. An example is ‘RADAR’. In terms of DNA, a palindrome is a sequence of base pairs that reads the same on both DNA strands when reading in the same orientation. For example:
5′ —— GAATTC —— 3′
3′ —— CTTAAG —— 5′
On reading both the strands shown above in the 5′ to 3′ direction, they give the same sequence. This is true even when they are both read in the 3′ to 5′ direction.
Restriction enzymes cut the DNA strand a little away from the center of the palindromic site, but between the same two bases on both strands. This gives rise to single-stranded, overhanging stretches on each strand called ‘sticky ends’. They are called ‘sticky’ because they can bind to their complementary cut counterparts.Using restriction enzymes we can generate recombinant DNA that contains DNA from different sources.
Cutting these DNA sources with the same restriction enzyme gives fragments with the same kind of ‘sticky ends’, that can then be joined using DNA ligases.
2) Cloning Vectors
Just the way a mosquito acts as a ‘vector’ to transfer the malarial parasite into the human body, we need vectors to transfer the cut DNA into a host organism. Vectors are one of the important tools of biotechnology.
Plasmids make good vectors because they can replicate in bacterial cells, independent of the control of the chromosomal DNA. The vectors in use currently are engineered such that they help in easy linking of foreign DNA and allow selection of recombinants over non-recombinants. A vector needs the following features to enable cloning:
(i) Origin of Replication (ori)
This is the sequence from where replication begins. Linking a piece of DNA to this sequence causes it to replicate in the host cell. This sequence also controls the copy number of the linked DNA. Therefore, the target DNA needs to be cloned into a vector whose ‘ori’ supports high copy number, in order to recover large amounts of the DNA.
(ii) Selectable Marker
The vector also needs to have a selectable marker which allows the selection of recombinants over non-recombinants. In terms of E. coli, some useful selectable markers are genes that provide resistance to antibiotics like ampicillin, kanamycin, chloramphenicol etc. Since the normal E. coli cells do not carry these resistance genes, it becomes easy to select the recombinants.
(iii) Cloning Sites
In order to attach the foreign DNA to a vector, the vector should have a recognition site for a specific restriction enzyme. Multiple recognition sites will result in multiple DNA fragments, complicating the process of cloning. A vector has more than one antibiotic resistance gene. The foreign DNA is ligated into a restriction site in one of the antibiotic resistance genes.
For example, let’s say an E. coli cloning vector has genes for ampicillin and tetracycline resistance. On ligating the foreign DNA into a recognition site within the tetracycline resistance gene, the plasmid loses its tetracycline resistance. But, it can still be selected for non-recombinants by plating on ampicillin-containing medium.
Now, by transferring the ones that grow in the ampicillin medium to a medium with tetracycline, we can dissect out recombinants from non-recombinants. The recombinants will grow in ampicillin but not in tetracycline medium; while non-recombinants will grow in both mediums.
Currently, alternative markers are available that can differentiate recombinants from non-recombinants based on their ability to produce color. This involves insertion of the foreign DNA in the DNA sequence of an enzyme like β-galactosidase, which inactivates the enzyme. This is ‘insertional inactivation’. On reaction with a substrate, the recombinants do not produce color whereas non-recombinants produce color.
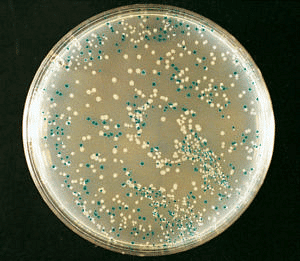 Blue White Screening
Blue White Screening
(iv) Vectors to Clone Genes in Plants and Animals
Long before us, bacteria and viruses knew how to transfer genes into plants and animals. For example, Agrobacterium tumifaciens, a pathogen on dicot plants, transfers ‘T-DNA’ that transforms normal plants cells into tumours. These tumours then produce chemicals that the pathogen requires. With better understanding, we have now converted these pathogens into useful vectors to deliver genes of interest to the plants or animals.
Now, the tumour-inducing (Ti) plasmid of Agrobacterium tumifaciens has been modified into a cloning vector. This vector is no longer harmful but is useful in delivering genes of interest to plants. Retroviruses transform normal cells into cancerous cells in animals. These have also been modified so that they are no longer harmful and can deliver genes to animals.
3) Competent Host
Host cells are bacterial cells which take up the recombinant DNA. Since DNA is hydrophilic, it cannot pass through the cell membrane of bacteria easily. Therefore, the bacterial cells have to be made ‘competent’ to take up the DNA.
Some procedures that make the cells competent are treatments with a specific concentration of divalent cation like calcium. This makes it easy for the DNA to enter the cell wall through pores. Incubation of cells with the recombinant DNA on the ice, followed by heat shock at 42°C and another incubation on ice, enables the cells to take up the DNA.
There are several other methods to introduce foreign DNA into host cells. The ‘microinjection’ method involves injecting the recombinant DNA directly into the nucleus of an animal cell. The ‘biolistics’ or ‘gene gun’ method bombards plant cells with high-velocity microparticles of gold or tungsten coated with DNA. The last method uses ‘disarmed pathogen vectors’ (discussed above) to transfer the recombinant DNA into the infected host cells.
|
59 videos|290 docs|168 tests
|
FAQs on Principles of Biotechnology: Genetic Engineering - Biology Class 12 - NEET
| 1. What is genetic engineering? |  |
| 2. What are the applications of genetic engineering? |  |
| 3. How is genetic engineering done? |  |
| 4. What are the ethical concerns associated with genetic engineering? |  |
| 5. What are the benefits of genetic engineering in medicine? |  |
















