NEET Exam > NEET Notes > Biology Class 12 > Important Diagrams: Molecular Basis of Inheritance
Important Diagrams: Molecular Basis of Inheritance | Biology Class 12 - NEET PDF Download
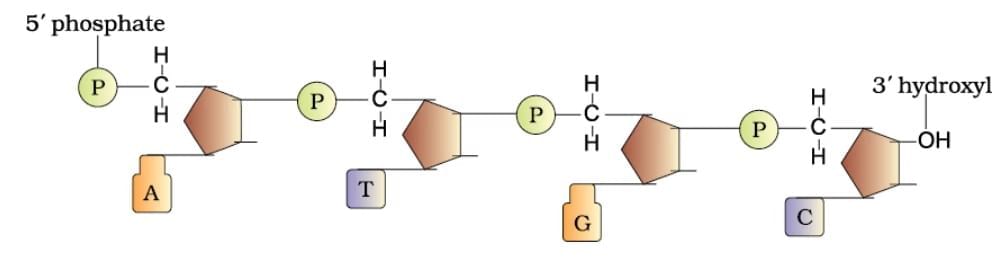
Polynucleotide Chain
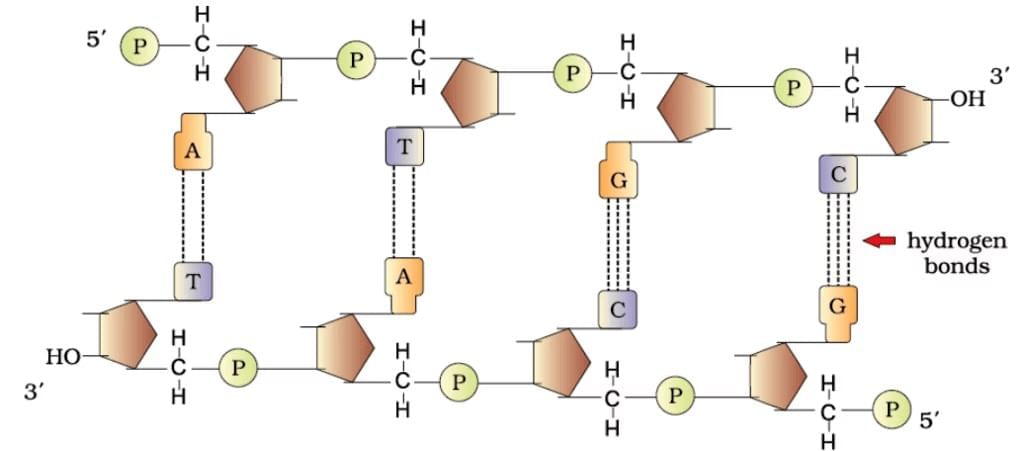
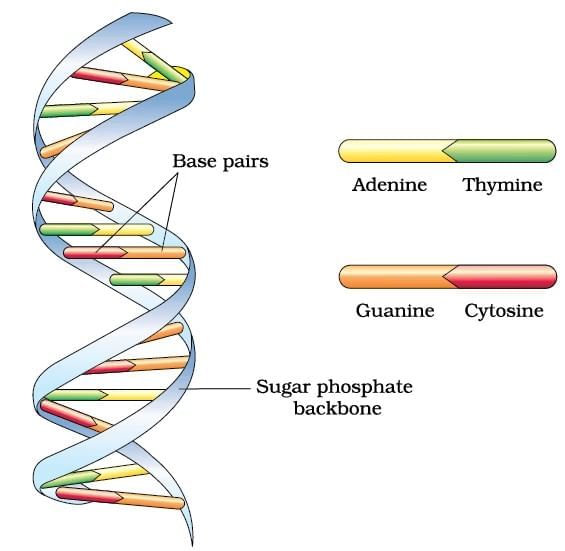
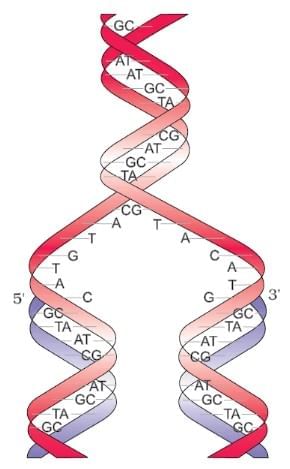
DNA Double Helix
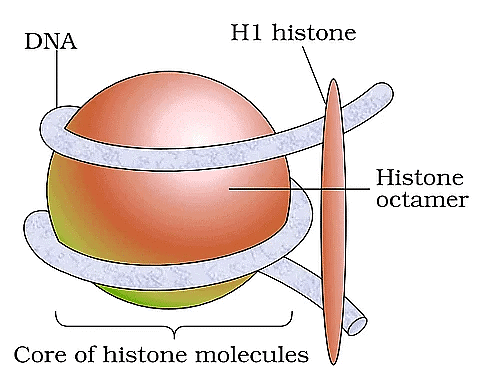
Nucleosome
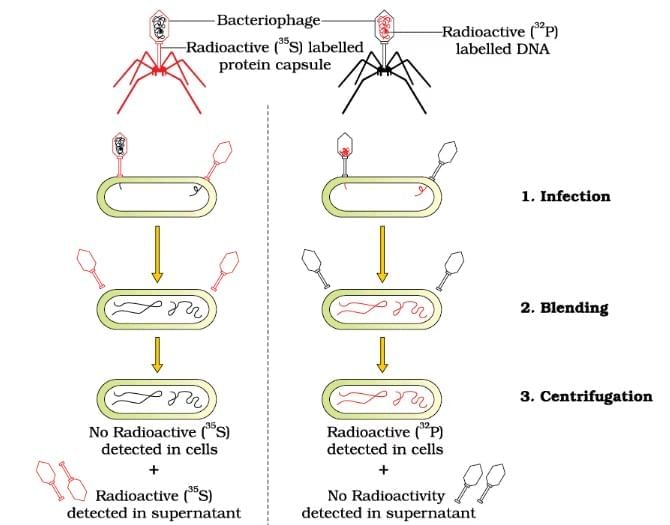
Hershey & Chase Experiment
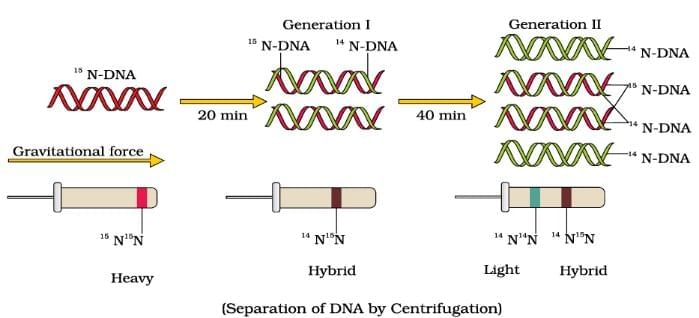
Semi Conservative DNA Replication ( Watson & Crick Model)
Meselson and Stahl’s Experiment
Replicating Fork
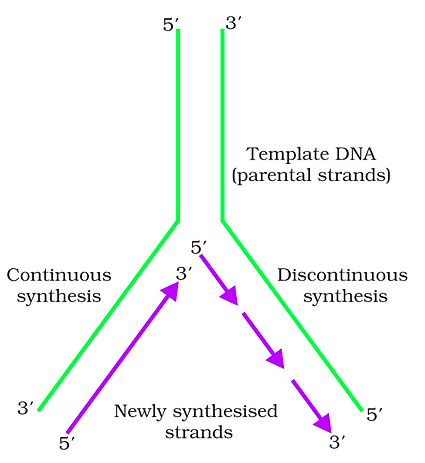
Transcription unit
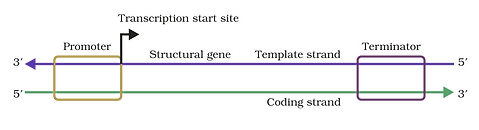
Transcription in Prokaryotes ( Bacteria)
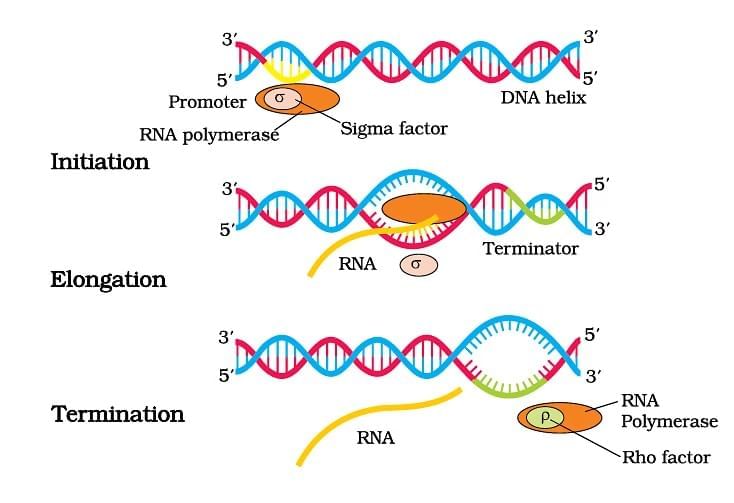
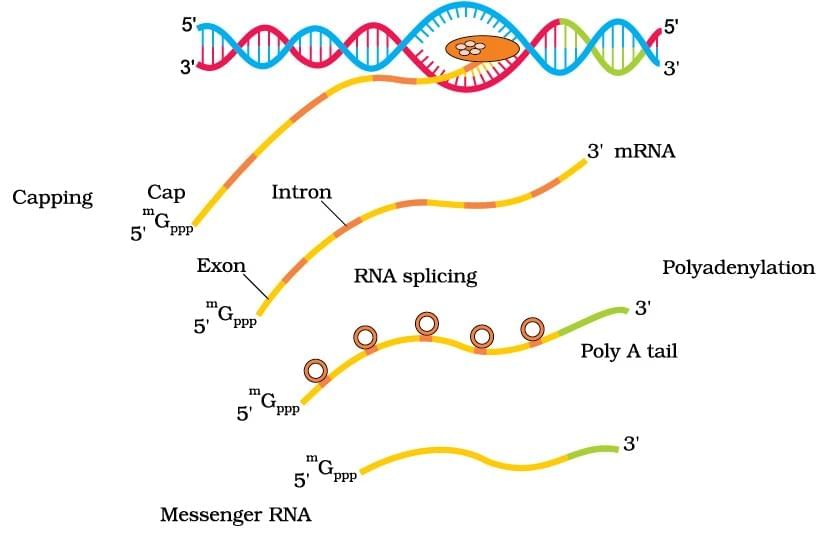
Transcription in Eukaryotes
Genetic Code
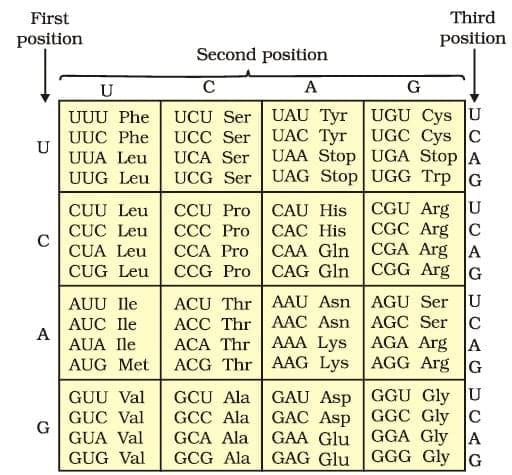
tRNA– the Adapter Molecule
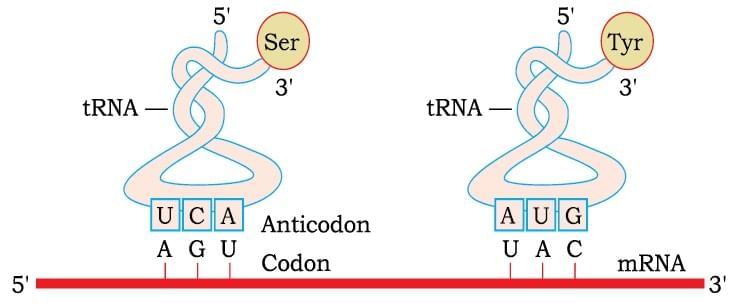
Translation
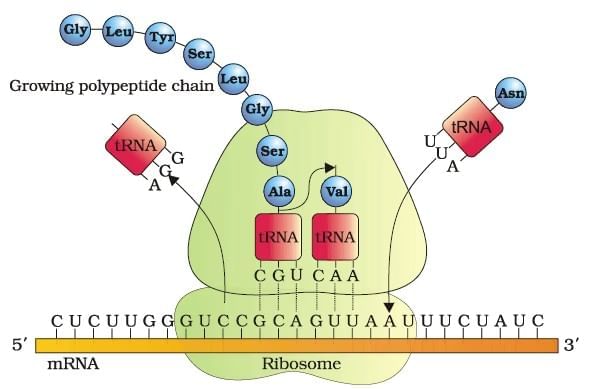
Lac operon
Human Genome Project
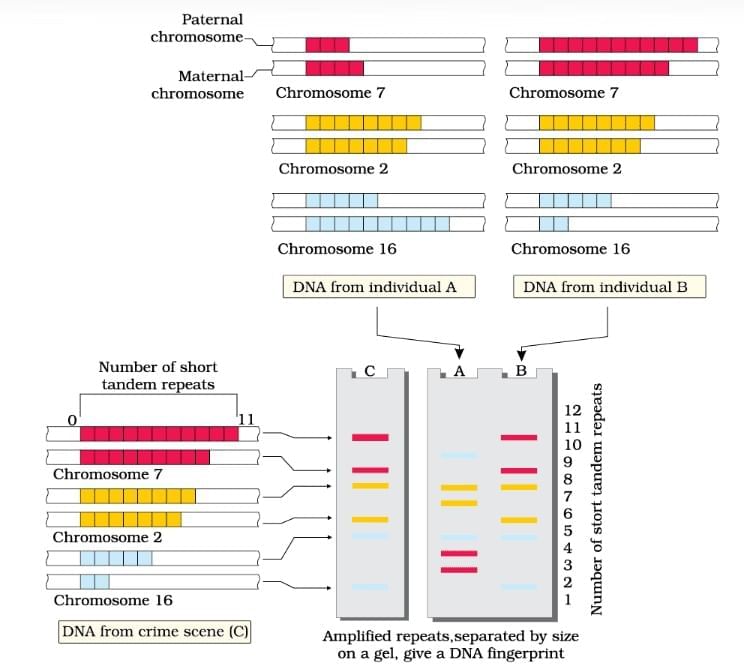
DNA Fingerprinting
The document Important Diagrams: Molecular Basis of Inheritance | Biology Class 12 - NEET is a part of the NEET Course Biology Class 12.
All you need of NEET at this link: NEET
|
59 videos|290 docs|168 tests
|
FAQs on Important Diagrams: Molecular Basis of Inheritance - Biology Class 12 - NEET
| 1. What is the structure of polynucleotide chains in DNA? |  |
Ans. Polynucleotide chains in DNA consist of long sequences of nucleotides, which are the building blocks of DNA. Each nucleotide is made up of three components: a phosphate group, a deoxyribose sugar, and a nitrogenous base (adenine, thymine, cytosine, or guanine). The nucleotides are linked together by phosphodiester bonds, forming a sugar-phosphate backbone, with the nitrogenous bases extending from the sugar. The two strands of DNA run in opposite directions (antiparallel) and twist to form a double helix.
| 2. How does DNA get packaged into a compact structure? |  |
Ans. DNA is packaged into a compact structure through a series of hierarchical levels of organization. Initially, DNA wraps around histone proteins to form nucleosomes, resembling "beads on a string." These nucleosomes further coil and fold into a more compact structure called chromatin. During cell division, chromatin condenses to form visible chromosomes, allowing for efficient segregation of genetic material.
| 3. What was the significance of the Hershey and Chase experiment? |  |
Ans. The Hershey and Chase experiment, conducted in 1952, provided crucial evidence that DNA is the genetic material in organisms. By using radioactive labeling, they tracked the DNA and protein components of bacteriophages (viruses that infect bacteria). Their results showed that only the viral DNA entered the bacterial cells and directed the production of new viruses, while the protein coat remained outside. This experiment confirmed that DNA carries genetic information.
| 4. What is semi-conservative DNA replication according to the Watson and Crick model? |  |
Ans. Semi-conservative DNA replication is a process where each of the two strands of the original DNA molecule serves as a template for the formation of new complementary strands. According to the Watson and Crick model, when DNA replicates, the double helix unwinds, and each strand is used to synthesize a new strand. As a result, each daughter DNA molecule consists of one old (parental) strand and one newly synthesized strand, preserving half of the original DNA.
| 5. What are the key differences in transcription between prokaryotes and eukaryotes? |  |
Ans. Transcription in prokaryotes occurs in the cytoplasm and involves a single RNA polymerase that synthesizes mRNA directly from the DNA template. In contrast, eukaryotic transcription takes place in the nucleus, where the primary mRNA undergoes processing (capping, polyadenylation, and splicing) before it is exported to the cytoplasm. Additionally, eukaryotic transcription involves multiple RNA polymerases and various transcription factors, whereas prokaryotes rely on simpler mechanisms.
Related Searches
















