RNA World, DNA Replication and its Enzymes | Biology Class 12 - NEET PDF Download
Introduction of RNA World
RNA was the first genetic material. There are evidences to prove that essential life processes, such as metabolism, translation, splicing, etc., have evolved around RNA.
(i) There are some important biochemical reactions in living systems that are catalysed by RNA catalysts and not by protein enzyme.
(ii) DNA has evolved from RNA with chemical modifications that make it more stable because RNA being a catalyst was reactive and hence, unstable.
There are following three types of RNAs:
(i) mRNA (messenger RNA) provides the template for transcription.
(ii) tRNA (transfer RNA) brings amino acids and reads the genetic code.
(iii) rRNA (ribosomal RNA) plays structural and catalytic role during translation.
All the three RNAs are needed to synthesise a protein in a cell.
Semiconservative Nature of DNA
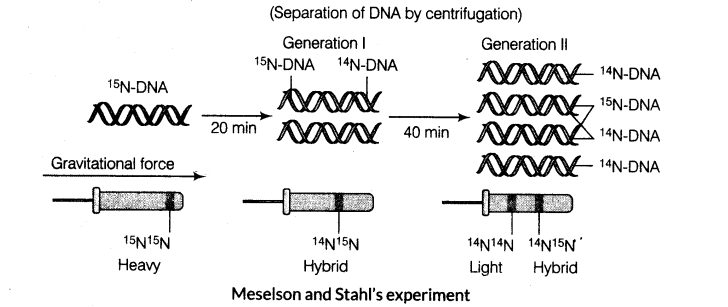
Matthew Meselson and Franklin Stahl Experiment
They performed the following experiments to prove this in 1958.
(i) E. coli was grown in a medium containing 15NH4Cl as the only nitrogen source for many generations. 15N got incorporated into newly synthesised DNA (and other nitrogen containing compounds). This heavy DNA molecule could be distinguished from the normal DNA by centrifugation in a cesium chloride (CsCl) density gradient.
(ii) They then transferred the cells into a medium with normal 14NH4Cl and took samples at various definite intervals as the cells multiplied and extracted the DNA that remained as double stranded helices. DNA samples were separated independently on CsCl gradients to measure DNA densities.
(iii) The DNA that was extracted from the culture, one generation (after 20 min) after the transfer from 15N to 14N medium had a hybrid or intermediate density. DNA extracted from the culture after another generation (after 40 min) was composed of equal amounts of this hybrid DNA and of light DNA.
(iv) Very similar experiments were carried out by Taylor and Colleagues on Vicia faba (faba beans) using radioactive thymidine and the same results, i.e. DNA replicates semi-conservatively, were obtained as in earlier experiments.
Introduction of DNA Replication
- At its most basic level, DNA replication is the operation of DNA polymerases producing a complementary DNA strand to the original template strand.
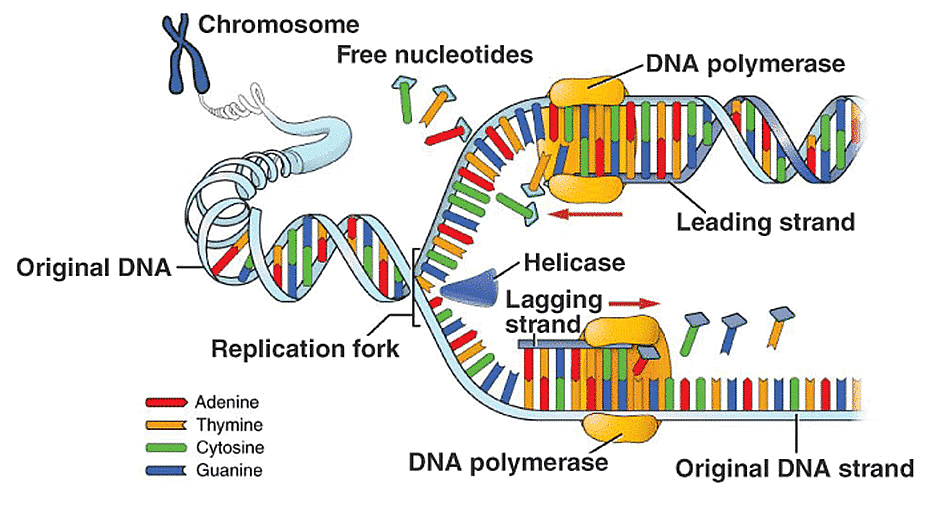
- To manufacture DNA, DNA helicases unwind double-stranded DNA before polymerases, generating a replication fork with two single-stranded templates.
- Replication allows a single DNA double helix to be copied into two DNA helices, which are then split into daughter cells during mitosis.
- From prokaryotes to eukaryotes, the primary enzymatic processes carried out at the replication fork are highly conserved.
- In actuality, the replication machinery is a large complex that coordinates several proteins at the replication site to create the “replisome.”
- Each proliferative cell’s replisome is in charge of duplicating the totality of genomic DNA.
- This mechanism is vital to all organisms because it provides for the high-fidelity transmission of hereditary/genetic information from parental cells to daughter cells.
DNA Replication Machinery
Factors involved in DNA replication and found on template ssDNAs makeup replication machineries.
- Primosotors include replication enzymes such as DNA polymerase, DNA helicases, DNA clamps, and DNA topoisomerases, as well as replication proteins such as single-stranded DNA binding proteins (SSB).
- These components work together in replication machines. All of the components involved in DNA replication are found on replication forks in most bacteria, and the complexes remain on the forks during DNA replication.
- Replisomes, or DNA replicase systems, are the replication machines. These words refer to proteins found on replication forks in general. Replisomes are not generated in eukaryotic and some bacterial cells.
- Replication factories are so named because replication machines do not move in relation to template DNAs like factories do.
- It’s a process that’s catalysed by enzymes. Artificial DNA primers and DNA polymerase, the principal enzyme in the replication process, can be employed to start DNA synthesis at known sequences in a template DNA molecule.
- DNA Replication Steps: Initiation, elongation, and termination are the three enzymatically catalysed and coordinated phases in DNA replication, as they are in other biological polymerisation processes.
- Initiation: DNA synthesis begins at specified locations along the DNA strand known as ‘origins,’ which include specific coding sequences. The origin of replication (ori) with all its regulatory elements make up the replicon. The ori is where DNA replication starts, allowing plasmids to self-replicate in order to live within cells. Initiator proteins target these origins, which then attract other proteins to enhance the replication process, producing a replication complex surrounding the DNA origin. Multiple origin sites occur inside the DNA structure; these locations are referred to as replication forks when DNA replication begins.The DNA helicase is found inside the replication complex. The double helix is unwound and each of the two strands is exposed, allowing them to be utilised as a template for reproduction. This is accomplished by hydrolysing the ATP needed to construct the nucleobase-to-nucleobase connections, therefore dissolving the link that holds the two strands together.
- Elongation: DNA Polymerase can begin synthesizing new strands of DNA to match the template strands once it has linked to the two unzipped strands of DNA (i.e., the template strands). Only free nucleotides can be added to the 3′ end of the primer by DNA polymerase.The new strand will be generated in a 5′ to 3′ direction since one of the template strands is read in a 3′ to 5′ direction. The leading strand refers to the freshly created strand. To launch DNA polymerase along the leading strand, DNA primase only has to synthesise an RNA primer once, at the start. This is due to the fact that DNA polymerase may lengthen the new DNA strand by reading the template 3′ to 5′ and synthesizing in the 5′ to 3′ direction, as mentioned previously.
- The lagging strand, on the other hand, is antiparallel and is read in a 5′ to 3′ direction. Continuous DNA synthesis in the 3′ to 5′ direction, as in the leading strand, would be impossible due to DNA polymerase’s inability to add nucleotides to the 5′ end. Instead, RNA primers are added to the newly exposed bases on the lagging strand as the helix unwinds, and DNA synthesis occurs in fragments, but still in the 5′ to 3′ orientation as before Termination: The process of extending new DNA strands continues until either no more DNA template strands can be replicated (i.e., at the chromosome’s end) or two replication forks meet and terminate. The meeting of two replication forks is uncontrolled and occurs at random throughout the chromosome’s length.The freshly synthesized strands are bound and stabilized when DNA synthesis is completed. Two enzymes are required to stabilize the lagging strand: RNAse H removes the RNA primer at the start of each Okazaki fragment, and DNA ligase binds the fragments together to form a single strand.
Enzymes Machinery and Enzymes of DNA Replication
Enzymes play an important role in DNA replication. DNA replication is aided by a variety of enzymes, including DNA-dependent DNA polymerase, helicase, and ligase.
- DNA helicase: Helix destabilizing enzyme is another name for it. At the Replication Fork, helicase divides the two DNA strands.
- DNA polymerase: During DNA replication, this enzyme catalyzes the addition of nucleotide substrates to DNA in the 5′ to 3′ orientation. In addition, he proofreads and corrects errors. DNA polymerase comes in a variety of forms, each of which serves a particular purpose in various types of cells.
- Single-stranded Binding Proteins: Bind to ssDNA to inhibit the DNA double helix from re-annealing after DNA helicase unwinds it, preserving strand separation and promoting strand synthesis.
- Topoisomerase: Relaxes the DNA, which is normally super-coiled.
- DNA gyrase: DNA helicase relieves the strain of unwinding; this is a special sort of topoisomerase.
- Primase: Provides a beginning place for DNA polymerase to initiate synthesis of the new DNA strand from RNA (or DNA).
- DNA Ligase: DNA ligase is a commercially available enzyme that is extracted from E.coli and Bacteriophage and is used in recombinant DNA technology. DNA ligase is an enzyme that combines DNA fragments using a cloning vector.
- Exonuclease: Exonucleases are a group of enzymes that cleave nucleotides from the 3′ or 5′ ends of DNA and RNA strands one at a time. Endonucleases, on the other hand, hydrolyze internal phosphodiester bonds, whereas this action does not.
What are the 2 Enzymes Used in DNA Replication…?
Unwinding of the template strand and polymerisation of the daughter strands are the two fundamental steps in DNA replication. As a result, the replicative helicase and polymerase are the two primary “workhorse” enzymes in the replisome.
- Replicative DNA Polymerases: With the help of other enzymes, it aids in polymerisation, catalyzes, and regulates the entire DNA replication process. The replication process uses deoxyribonucleoside triphosphates as both a substrate and an energy source.
- Replicative DNA Helicases: The double-stranded helix must be unfolded to disclose a single-stranded template for DNA polymerases to operate. The replicative helicase is in charge of this task. The replicative helicase in eukaryotes is a hexameric complex made up of minichromosome maintenance proteins.
Conclusion
Factors involved in DNA replication and found on template ssDNAs makeup replication machineries. Promoters include replication enzymes such as DNA polymerase, DNA helicases, DNA clamps, and DNA topoisomerases, as well as replication proteins such as single-stranded DNA binding proteins (SSB). These components work together in replication machines. All of the components involved in DNA replication are found on replication forks in most bacteria, and the complexes remain on the forks during DNA replication. Replisomes, or DNA replicase systems, are the replication machines.
|
59 videos|290 docs|168 tests
|
FAQs on RNA World, DNA Replication and its Enzymes - Biology Class 12 - NEET
| 1. What is the RNA World hypothesis? |  |
| 2. How does the DNA replication machinery work? |  |
| 3. What are the key enzymes involved in DNA replication? |  |
| 4. What was the Meselson-Stahl experiment? |  |
| 5. How does the RNA world hypothesis relate to DNA replication? |  |

















