Regulation of Gene Expression | Biology Class 12 - NEET PDF Download
| Table of contents |

|
| Regulation of Gene Expression |

|
| The Lac Operon |

|
| Human Genome Project |

|
| DNA Fingerprinting |

|
Regulation of Gene Expression
Gene expression regulation is a broad term that can occur at various levels. Since gene expression leads to the formation of a polypeptide, it can be regulated at several stages.
In eukaryotes, regulation can happen at:
- Transcriptional level: Involves the formation of the primary transcript.
- Processing level: Regulation of splicing.
- Transport: Movement of mRNA from the nucleus to the cytoplasm.
- Translational level: Control over the translation process.
Genes within a cell are expressed to perform specific functions. For example, when the bacterium E. coli produces the enzyme beta-galactosidase, it helps break down the disaccharide lactose into galactose and glucose, which the bacteria use for energy. If lactose is not available, the bacteria no longer need to produce this enzyme. Thus, gene expression is regulated by metabolic, physiological, or environmental conditions.
In prokaryotes,
the control of transcriptional initiation is the primary means of regulating gene expression. The activity of RNA polymerase at a promoter is influenced by accessory proteins that help it recognize start sites. These regulatory proteins can act as activators (positive control) or repressors (negative control).
The accessibility of promoter regions in prokaryotic DNA is often regulated by proteins interacting with sequences called operators. The operator region, usually located next to the promoter, binds repressor proteins specific to each operon. For instance, the lac operator is unique to the lac operon and interacts only with the lac repressor.
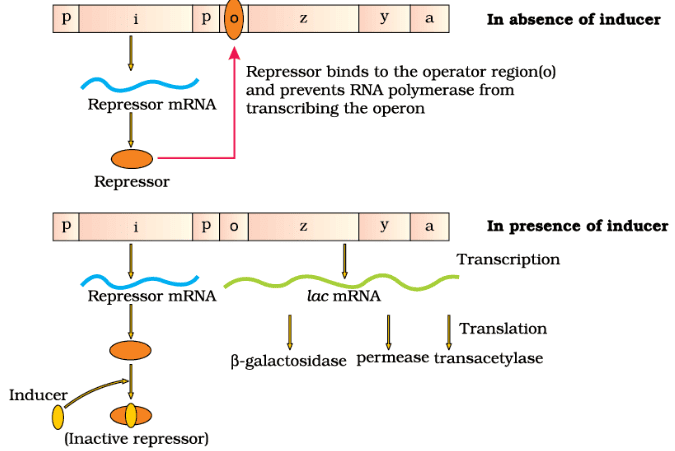
The Lac Operon
The lac operon was elucidated by Francois Jacob and Jacque Monod, who studied a transcriptionally regulated system in bacteria. The lac operon, involving the metabolism of lactose, is a classic example of gene regulation in prokaryotes.
Key Components of the Lac Operon:
(i) Regulatory Gene (i gene): This gene codes for the repressor protein that inhibits the operon.
(ii) Structural Genes: The lac operon comprises three structural genes:
- z Gene: Codes for beta-galactosidase, which hydrolyzes lactose into glucose and galactose.
- y Gene: Codes for permease, which facilitates the entry of lactose into the bacterial cell.
- a Gene: Codes for transacetylase, involved in lactose metabolism.
 The Lac Operon
The Lac Operon
Regulation of the Lac Operon:
- Induction: Lactose acts as an inducer by binding to the repressor and inactivating it. This allows RNA polymerase to access the promoter and initiate transcription.
- Negative Regulation: The lac operon is primarily regulated by the repressor protein, which prevents transcription when lactose is absent.
- Constitutive Expression: The repressor is synthesized continuously from the i gene, ensuring tight regulation of the operon.
Operons in Bacteria:
Similar to the lac operon, other operons in bacteria, such as the trp operon, ara operon, his operon, and val operon, consist of structural genes regulated by a common promoter and regulatory elements. These operons enable bacteria to efficiently regulate gene expression in response to environmental changes and metabolic needs.
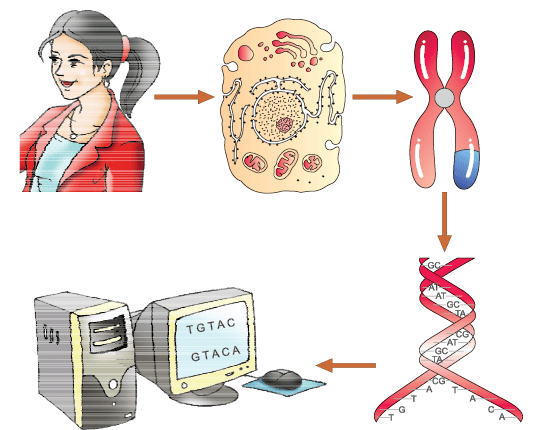
Human Genome Project
The Human Genome Project (HGP) was an ambitious scientific endeavor aimed at mapping and understanding all the genes of the human species. Here's a simplified overview:
What is DNA and Why is it Important?
- DNA is like a blueprint for building and operating a living organism. It contains the genetic instructions that determine everything from eye color to the risk of certain diseases.
- The sequence of bases (A, T, C, and G) in DNA is what makes each individual's genetic code unique. Even small differences in these sequences can lead to different traits or health conditions.
What Did the HGP Aim to Do?
- The HGP aimed to read and map the entire human DNA sequence, which is about 3 billion base pairs long.
- It set out to identify all the estimated 20,000-25,000 genes in human DNA and determine the order of the 3 billion chemical base pairs that make up human DNA.
- The project also aimed to store this vast amount of information in databases, improve data analysis tools, and address ethical, legal, and social issues related to genetic research.
Why was the HGP Considered a Mega Project?
- The sheer scale of the project was enormous. With the initial cost estimated at about 9 billion US dollars and the need for vast amounts of data storage and analysis, the HGP was a massive undertaking.
- If the DNA sequences were printed, they would require thousands of books to store all the information from just one human cell!
- The HGP also required advanced computer technology for data storage, retrieval, and analysis, leading to the growth of a new field called Bioinformatics.
How Was the HGP Accomplished?
- The project, which lasted 13 years, was coordinated by the U.S. Department of Energy and the National Institute of Health, with significant contributions from the Wellcome Trust in the U.K. and other countries like Japan, France, Germany, and China.
- It was completed in 2003, successfully mapping the human genome and providing invaluable insights into human biology and health.
What Were the Benefits of the HGP?
- The knowledge gained from the HGP has the potential to revolutionize medicine by improving the diagnosis and treatment of diseases.
- Understanding DNA variations among individuals can lead to new ways to prevent and treat various disorders.
- Research on non-human DNA sequences can also help in areas like agriculture, energy production, and environmental cleanup.
Methodologies
Two Major Approaches:
- Expressed Sequence Tags (ESTs): This approach focused on identifying all the genes that are expressed as RNA.
- Sequence Annotation: This approach involved sequencing the entire genome, including both coding and non-coding sequences, and later assigning functions to different regions.
Sequencing Process
- DNA Isolation and Fragmentation: The total DNA from a cell is isolated and converted into random fragments of smaller sizes due to technical limitations in sequencing long pieces of DNA.
- Cloning: The DNA fragments are cloned into suitable hosts using specialized vectors, resulting in the amplification of each fragment for easier sequencing.
- Common Hosts and Vectors: The commonly used hosts are bacteria and yeast, with vectors such as BAC (bacterial artificial chromosomes) and YAC (yeast artificial chromosomes).
- Sequencing Method: The fragments are sequenced using automated DNA sequencers based on the method developed by Frederick Sanger.
Sequence Assembly and Annotation
- The sequenced fragments are arranged based on overlapping regions.
- Specialized computer programs are developed to align these sequences due to the complexity of the task.
- The sequences are then annotated and assigned to each chromosome.
Completion of Chromosome Sequencing
The sequence of chromosome 1 was completed in May 2006, marking the completion of all 24 human chromosomes (22 autosomes and X and Y).
Genetic and Physical Mapping
The genetic and physical maps of the genome were generated using information on polymorphism of restriction endonuclease recognition sites and repetitive DNA sequences known as microsatellites.
Salient Features of Human Genome
- The human genome comprises approximately 3.164 billion base pairs (bp) of DNA.
- The average gene in the human genome is about 3,000 bases long, but there is significant variation in gene size. For instance, the largest known human gene, dystrophin, spans about 2.4 million bases.
- Current estimates suggest that there are around 30,000 genes in the human genome, which is much lower than earlier estimates ranging from 80,000 to 140,000 genes.
- Almost all (99.9%) nucleotide bases are identical in all humans, indicating a high level of genetic similarity across individuals.
- More than 50% of the identified genes have unknown functions, highlighting gaps in our understanding of the genome.
- Less than 2% of the genome is involved in coding for proteins, indicating that a significant portion of the DNA does not have a direct role in protein synthesis.
- Repeated sequences constitute a large part of the human genome. These are stretches of DNA that are repeated many times, sometimes hundreds or thousands of times.
- Chromosome 1 contains the most genes (2,968), while the Y chromosome has the fewest (231).
- Scientists have identified approximately 1.4 million locations in the genome where single-base DNA differences, known as single nucleotide polymorphisms (SNPs), occur. This information is valuable for understanding genetic variation and its implications for health and disease.
Applications and Future Challenges
- Deriving meaningful knowledge from DNA sequences will define research in the coming decades, enhancing our understanding of biological systems.
- This task will require the expertise and creativity of thousands of scientists from various disciplines globally.
- The Human Genome (HG) sequence enables a new approach to biological research, allowing scientists to study all genes in a genome systematically.
- Researchers can now investigate gene networks, transcripts, and how genes and proteins interact on a much broader scale than before.
DNA Fingerprinting
DNA fingerprinting is a powerful technique used to identify genetic differences between individuals. It focuses on specific regions of the DNA that show high variability among people. Here are the key points:
Understanding Genetic Differences
- Humans share 99.9% of their DNA sequence, but the tiny differences in the remaining 0.1% make each person unique.
- With a human genome of about 3 billion base pairs (bp), the differences are found in approximately 3 million base sequences.
What is DNA Fingerprinting?
- DNA fingerprinting is a quick and efficient method to compare DNA sequences from different individuals.
- It focuses on specific regions of DNA known as repetitive DNA, where short stretches of DNA are repeated many times.
How Does It Work?
- During a process called density gradient centrifugation, repetitive DNA separates into different peaks based on its composition and length.
- The main peak represents the bulk DNA, while smaller peaks, called satellite DNA, represent the repetitive regions.
- These regions are classified into categories like micro-satellites and mini-satellites.
Why is it Useful?
- Repetitive DNA does not code for proteins but varies greatly among individuals, making it ideal for identification.
- DNA from different tissues, such as blood, hair, or saliva, shows the same degree of variability, making it a reliable tool for forensic applications.
- DNA fingerprinting is also used in paternity testing to establish familial relationships.
What is DNA Polymorphism?
- DNA polymorphism refers to variations in the DNA sequence that arise due to mutations.
- Mutations can occur in somatic cells or germ cells (cells that produce gametes).
- If a mutation in a germ cell does not hinder reproduction, it can spread through the population.
- DNA polymorphism is identified when a variant occurs in a population with a frequency greater than 1%.
Types of Polymorphisms
- Polymorphisms can range from single nucleotide changes to large-scale alterations in the DNA.
- These variations play a crucial role in evolution and speciation.
Development of DNA Fingerprinting
- The technique of DNA fingerprinting was pioneered by Alec Jeffreys, who used satellite DNA showing high polymorphism.
- This involved a process called Variable Number of Tandem Repeats (VNTR), where the number of repeats in a DNA sequence varies between individuals.
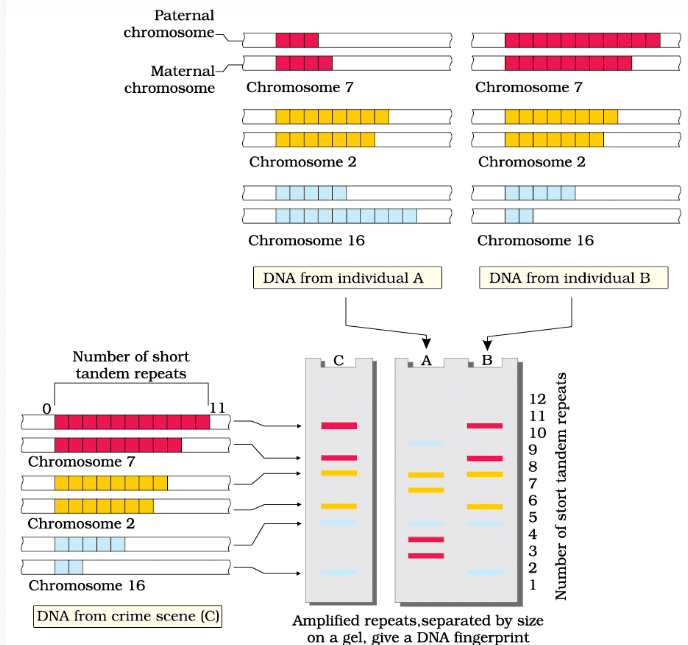
Steps in DNA Fingerprinting
- Isolation of DNA: DNA is extracted from the sample.
- Digestion: DNA is cut into fragments using restriction enzymes.
- Separation: Fragments are separated by size through gel electrophoresis.
- Blotting: DNA fragments are transferred to a membrane.
- Hybridization: Membrane is probed with labeled VNTR sequences.
- Detection: Hybridized fragments are visualized, creating a unique pattern for each individual.
Advancements in DNA Fingerprinting
- The sensitivity of the technique has improved with the use of polymerase chain reaction (PCR), allowing analysis from a single cell.
- Besides forensic applications, DNA fingerprinting is used in studying population and genetic diversities.
- Different probes are now available to generate DNA fingerprints, enhancing the technique's versatility.
|
59 videos|290 docs|168 tests
|
FAQs on Regulation of Gene Expression - Biology Class 12 - NEET
| 1. What is the Lac Operon and how does it function in prokaryotic gene regulation? |  |
| 2. What role does the repressor play in the regulation of the Lac Operon? |  |
| 3. How does glucose influence the regulation of the Lac Operon? |  |
| 4. What are the structural genes of the Lac Operon and their functions? |  |
| 5. What happens to the Lac Operon when both lactose and glucose are present? |  |




















