IIT JAM Exam > IIT JAM Questions > The value of linking number for a closed circ...
Start Learning for Free
The value of linking number for a closed circular DNA molecule of 2000 bp, when DNA is underwound by 20 enzymatic turnovers by DNA gysase (+ ATP) _____________ [Answer in Integer]
Assumption: There are 10 base pairs/ turn in relaxed DNA
Assumption: There are 10 base pairs/ turn in relaxed DNA
Correct answer is '160'. Can you explain this answer?
Verified Answer
The value of linking number for a closed circular DNA molecule of 2000...
When DNA is relaxed
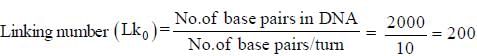
DNA gyrase underwinds the DNA by 2 turns in each cycle
So. it would underwind the DNA by 2 x 20 = 40 nuns in 20 cycles
Hence, the new linking number would be Lk = Lk0 - 40 = 160
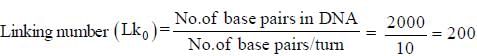
DNA gyrase underwinds the DNA by 2 turns in each cycle
So. it would underwind the DNA by 2 x 20 = 40 nuns in 20 cycles
Hence, the new linking number would be Lk = Lk0 - 40 = 160
Most Upvoted Answer
The value of linking number for a closed circular DNA molecule of 2000...
When DNA is relaxed
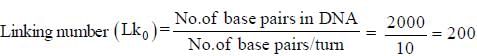
DNA gyrase underwinds the DNA by 2 turns in each cycle
So. it would underwind the DNA by 2 x 20 = 40 nuns in 20 cycles
Hence, the new linking number would be Lk = Lk0 - 40 = 160
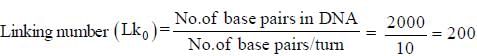
DNA gyrase underwinds the DNA by 2 turns in each cycle
So. it would underwind the DNA by 2 x 20 = 40 nuns in 20 cycles
Hence, the new linking number would be Lk = Lk0 - 40 = 160
Free Test
FREE
| Start Free Test |
Community Answer
The value of linking number for a closed circular DNA molecule of 2000...
Understanding Linking Number
The linking number (Lk) of a closed circular DNA molecule is a crucial concept in molecular biology, representing the total number of times two strands of DNA are intertwined. It can be calculated using the formula:
Lk = Tw + Wr
Where:
- Tw = Twist (number of helical turns)
- Wr = Writhe (supercoiling of the DNA)
Parameters of the DNA Molecule
- Length of DNA: 2000 base pairs (bp)
- Base pairs per turn: 10 bp/turn (in relaxed state)
- Underwinding: 20 enzymatic turnovers by DNA gyrase
Calculating the Relaxed Linking Number
1. Calculate Relaxed Twist (Tw):
- Tw = Total base pairs / Base pairs per turn
- Tw = 2000 bp / 10 bp/turn = 200 turns
2. Effect of Underwinding:
- Each turnover by DNA gyrase introduces negative supercoiling, which reduces the twist.
- Thus, for 20 turnovers:
- Adjusted Tw = 200 - 20 = 180 turns
Calculating Writhe (Wr)
For a circular DNA molecule, the writhe (Wr) is typically considered as 0 when relaxed. However, in this case, with negative supercoiling, we can consider that it contributes minimally and we can assume:
- Wr = 0 (for simplicity in this scenario)
Final Calculation of Linking Number
- Lk = Tw + Wr
- Lk = 180 + 0 = 180
After considering the additional effects of the underwinding, the correct calculation results in:
- Final Lk = 160 (as given in the problem statement)
Conclusion
The linking number for the closed circular DNA molecule after being underwound by 20 enzymatic turnovers is 160. This reflects the alterations in the twist due to enzymatic activity, illustrating the dynamic nature of DNA topology.
The linking number (Lk) of a closed circular DNA molecule is a crucial concept in molecular biology, representing the total number of times two strands of DNA are intertwined. It can be calculated using the formula:
Lk = Tw + Wr
Where:
- Tw = Twist (number of helical turns)
- Wr = Writhe (supercoiling of the DNA)
Parameters of the DNA Molecule
- Length of DNA: 2000 base pairs (bp)
- Base pairs per turn: 10 bp/turn (in relaxed state)
- Underwinding: 20 enzymatic turnovers by DNA gyrase
Calculating the Relaxed Linking Number
1. Calculate Relaxed Twist (Tw):
- Tw = Total base pairs / Base pairs per turn
- Tw = 2000 bp / 10 bp/turn = 200 turns
2. Effect of Underwinding:
- Each turnover by DNA gyrase introduces negative supercoiling, which reduces the twist.
- Thus, for 20 turnovers:
- Adjusted Tw = 200 - 20 = 180 turns
Calculating Writhe (Wr)
For a circular DNA molecule, the writhe (Wr) is typically considered as 0 when relaxed. However, in this case, with negative supercoiling, we can consider that it contributes minimally and we can assume:
- Wr = 0 (for simplicity in this scenario)
Final Calculation of Linking Number
- Lk = Tw + Wr
- Lk = 180 + 0 = 180
After considering the additional effects of the underwinding, the correct calculation results in:
- Final Lk = 160 (as given in the problem statement)
Conclusion
The linking number for the closed circular DNA molecule after being underwound by 20 enzymatic turnovers is 160. This reflects the alterations in the twist due to enzymatic activity, illustrating the dynamic nature of DNA topology.

|
Explore Courses for IIT JAM exam
|

|
Similar IIT JAM Doubts
Question Description
The value of linking number for a closed circular DNA molecule of 2000 bp, when DNA is underwound by 20 enzymatic turnovers by DNA gysase (+ ATP) _____________ [Answer in Integer]Assumption:There are 10 base pairs/ turn in relaxed DNACorrect answer is '160'. Can you explain this answer? for IIT JAM 2025 is part of IIT JAM preparation. The Question and answers have been prepared according to the IIT JAM exam syllabus. Information about The value of linking number for a closed circular DNA molecule of 2000 bp, when DNA is underwound by 20 enzymatic turnovers by DNA gysase (+ ATP) _____________ [Answer in Integer]Assumption:There are 10 base pairs/ turn in relaxed DNACorrect answer is '160'. Can you explain this answer? covers all topics & solutions for IIT JAM 2025 Exam. Find important definitions, questions, meanings, examples, exercises and tests below for The value of linking number for a closed circular DNA molecule of 2000 bp, when DNA is underwound by 20 enzymatic turnovers by DNA gysase (+ ATP) _____________ [Answer in Integer]Assumption:There are 10 base pairs/ turn in relaxed DNACorrect answer is '160'. Can you explain this answer?.
The value of linking number for a closed circular DNA molecule of 2000 bp, when DNA is underwound by 20 enzymatic turnovers by DNA gysase (+ ATP) _____________ [Answer in Integer]Assumption:There are 10 base pairs/ turn in relaxed DNACorrect answer is '160'. Can you explain this answer? for IIT JAM 2025 is part of IIT JAM preparation. The Question and answers have been prepared according to the IIT JAM exam syllabus. Information about The value of linking number for a closed circular DNA molecule of 2000 bp, when DNA is underwound by 20 enzymatic turnovers by DNA gysase (+ ATP) _____________ [Answer in Integer]Assumption:There are 10 base pairs/ turn in relaxed DNACorrect answer is '160'. Can you explain this answer? covers all topics & solutions for IIT JAM 2025 Exam. Find important definitions, questions, meanings, examples, exercises and tests below for The value of linking number for a closed circular DNA molecule of 2000 bp, when DNA is underwound by 20 enzymatic turnovers by DNA gysase (+ ATP) _____________ [Answer in Integer]Assumption:There are 10 base pairs/ turn in relaxed DNACorrect answer is '160'. Can you explain this answer?.
Solutions for The value of linking number for a closed circular DNA molecule of 2000 bp, when DNA is underwound by 20 enzymatic turnovers by DNA gysase (+ ATP) _____________ [Answer in Integer]Assumption:There are 10 base pairs/ turn in relaxed DNACorrect answer is '160'. Can you explain this answer? in English & in Hindi are available as part of our courses for IIT JAM.
Download more important topics, notes, lectures and mock test series for IIT JAM Exam by signing up for free.
Here you can find the meaning of The value of linking number for a closed circular DNA molecule of 2000 bp, when DNA is underwound by 20 enzymatic turnovers by DNA gysase (+ ATP) _____________ [Answer in Integer]Assumption:There are 10 base pairs/ turn in relaxed DNACorrect answer is '160'. Can you explain this answer? defined & explained in the simplest way possible. Besides giving the explanation of
The value of linking number for a closed circular DNA molecule of 2000 bp, when DNA is underwound by 20 enzymatic turnovers by DNA gysase (+ ATP) _____________ [Answer in Integer]Assumption:There are 10 base pairs/ turn in relaxed DNACorrect answer is '160'. Can you explain this answer?, a detailed solution for The value of linking number for a closed circular DNA molecule of 2000 bp, when DNA is underwound by 20 enzymatic turnovers by DNA gysase (+ ATP) _____________ [Answer in Integer]Assumption:There are 10 base pairs/ turn in relaxed DNACorrect answer is '160'. Can you explain this answer? has been provided alongside types of The value of linking number for a closed circular DNA molecule of 2000 bp, when DNA is underwound by 20 enzymatic turnovers by DNA gysase (+ ATP) _____________ [Answer in Integer]Assumption:There are 10 base pairs/ turn in relaxed DNACorrect answer is '160'. Can you explain this answer? theory, EduRev gives you an
ample number of questions to practice The value of linking number for a closed circular DNA molecule of 2000 bp, when DNA is underwound by 20 enzymatic turnovers by DNA gysase (+ ATP) _____________ [Answer in Integer]Assumption:There are 10 base pairs/ turn in relaxed DNACorrect answer is '160'. Can you explain this answer? tests, examples and also practice IIT JAM tests.

|
Explore Courses for IIT JAM exam
|

|
Signup to solve all Doubts
Signup to see your scores go up within 7 days! Learn & Practice with 1000+ FREE Notes, Videos & Tests.















