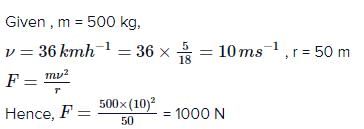All Exams >
IIT JAM >
IIT JAM Past Year Papers and Model Test Paper (All Branches) >
All Questions
All questions of Biotechnology - BT for IIT JAM Exam
Match the entries in Group I with the entries in Group II
- a)P-iv, Q-i, R-ii, S-iii
- b)P-iv, Q-i, R-iii, S-ii
- c)P-iv, Q-iii, R-ii, S-i
- d)P-ii, Q-iv, R-i, S-iii
Correct answer is option 'A'. Can you explain this answer?
Match the entries in Group I with the entries in Group II

a)
P-iv, Q-i, R-ii, S-iii
b)
P-iv, Q-i, R-iii, S-ii
c)
P-iv, Q-iii, R-ii, S-i
d)
P-ii, Q-iv, R-i, S-iii

|
Veda Institute answered |
Correct Answer :- a
Explanation : a) Nylon -6,6 is synthesized by polycondensation of hexamethylenediamine and adipic acid.
b) Natural rubber consists mainly of poly-cis-isoprene with a molecular mass of 100,000 to 1,000,000 g/mol. Some natural rubber sources, called gutta percha, are composed of trans-1,4-polyisoprene, a structural isomer that has similar, but not identical, properties.
c) Of the hexoses, glucose, fructose, and galactose are physiologically the most important. Glucose is a major mammalian fuel, found widely in fruits and vegetables as a monosaccharide, in disaccharides such as sucrose, maltose, and lactose, and in polysaccharides such as glycogen and starch.
d) Myoglobin is a small 153-amino acid protein encoded by the MB gene and is the primary carrier and storage center of oxygen in muscle.
Which one is not correct about perxenate ion (XeO64–) ?- a)Hydrolysis of XeF6 in basic medium gives perxenate ion
- b)It has two oxygen involved in peroxide bond
- c)Oxidation number of xenon in Perxenate ion is +8
- d)Perxenates are Strong oxidizing agents.
Correct answer is option 'B'. Can you explain this answer?
Which one is not correct about perxenate ion (XeO64–) ?
a)
Hydrolysis of XeF6 in basic medium gives perxenate ion
b)
It has two oxygen involved in peroxide bond
c)
Oxidation number of xenon in Perxenate ion is +8
d)
Perxenates are Strong oxidizing agents.

|
Bijoy Patel answered |
Explanation:
Perxenate ion (XeO64) is a polyatomic ion formed by the reaction of XeF6 with a strong base. Let's discuss the given options one by one.
a) Hydrolysis of XeF6 in basic medium gives perxenate ion:
This statement is correct. XeF6 is a fluoride of xenon and reacts with a strong base like NaOH or KOH to form perxenate ion. The reaction is given below:
XeF6 + 6 NaOH → Na4XeO6 + 2 NaF + 3 H2O
b) It has two oxygen involved in peroxide bond:
This statement is incorrect. Perxenate ion does not have a peroxide bond. It has four oxygen atoms bonded to xenon in a square planar arrangement.
c) Oxidation number of xenon in Perxenate ion is 8:
This statement is correct. The oxidation number of xenon in perxenate ion is +8. Each oxygen atom has an oxidation number of -2, and since there are six oxygen atoms, the total oxidation number contributed by oxygen is -12. Therefore, the oxidation number of xenon is +8 to balance the charge of the ion.
d) Perxenates are Strong oxidizing agents:
This statement is correct. Perxenates are strong oxidizing agents and can oxidize a wide range of compounds. For example, they can oxidize iodide ion to iodine, sulfur dioxide to sulfate ion, and ferrous ion to ferric ion.
In conclusion, statement b is incorrect as perxenate ion does not have a peroxide bond.
Perxenate ion (XeO64) is a polyatomic ion formed by the reaction of XeF6 with a strong base. Let's discuss the given options one by one.
a) Hydrolysis of XeF6 in basic medium gives perxenate ion:
This statement is correct. XeF6 is a fluoride of xenon and reacts with a strong base like NaOH or KOH to form perxenate ion. The reaction is given below:
XeF6 + 6 NaOH → Na4XeO6 + 2 NaF + 3 H2O
b) It has two oxygen involved in peroxide bond:
This statement is incorrect. Perxenate ion does not have a peroxide bond. It has four oxygen atoms bonded to xenon in a square planar arrangement.
c) Oxidation number of xenon in Perxenate ion is 8:
This statement is correct. The oxidation number of xenon in perxenate ion is +8. Each oxygen atom has an oxidation number of -2, and since there are six oxygen atoms, the total oxidation number contributed by oxygen is -12. Therefore, the oxidation number of xenon is +8 to balance the charge of the ion.
d) Perxenates are Strong oxidizing agents:
This statement is correct. Perxenates are strong oxidizing agents and can oxidize a wide range of compounds. For example, they can oxidize iodide ion to iodine, sulfur dioxide to sulfate ion, and ferrous ion to ferric ion.
In conclusion, statement b is incorrect as perxenate ion does not have a peroxide bond.
Kinetic theory of an ideal gas is based upon the following assumption(s)- a)Gases are made of molecules with negligible volume
- b)The gaseous molecules do not possess kinetic energy
- c)The molecules are in constant random motion
- d)Intermolecular forces of attraction are negligible
Correct answer is option 'A,C,D'. Can you explain this answer?
Kinetic theory of an ideal gas is based upon the following assumption(s)
a)
Gases are made of molecules with negligible volume
b)
The gaseous molecules do not possess kinetic energy
c)
The molecules are in constant random motion
d)
Intermolecular forces of attraction are negligible
|
|
Arghya answered |
If u go through the postulates of Kinetic theory of ideal gases then it will be clear to you.
for a real gas (p+an^2/v^2)(v-nb) , where b is the excluded volume and a= intermolecular force of attraction.
but for an ideal gas the equn is PV = nRT.
thus , A,C,D are correct...
for a real gas (p+an^2/v^2)(v-nb) , where b is the excluded volume and a= intermolecular force of attraction.
but for an ideal gas the equn is PV = nRT.
thus , A,C,D are correct...
At what pH does poly-Glu in an aqueous solution form α-helical structure?- a)3
- b)7
- c)9
- d)12
Correct answer is option 'A'. Can you explain this answer?
At what pH does poly-Glu in an aqueous solution form α-helical structure?
a)
3
b)
7
c)
9
d)
12

|
Shubham Rane answered |
Poly-Glu can form in an aqueous solution at any pH, as it is not affected by changes in pH. However, the solubility and conformation of the poly-Glu molecules may be affected by the pH of the solution. At low pH, the poly-Glu may become more compact and less soluble, while at high pH, it may become more extended and more soluble.
Two dice are thrown simultaneously. The probability that the sum of the number obtained is divisible by 7 is- a)1/6
- b)1/36
- c)0
- d)1/18
Correct answer is option 'A'. Can you explain this answer?
Two dice are thrown simultaneously. The probability that the sum of the number obtained is divisible by 7 is
a)
1/6
b)
1/36
c)
0
d)
1/18

|
Chirag Nambiar answered |
Solution:
When two dice are thrown simultaneously, the total number of outcomes = 6 x 6 = 36.
We need to find the probability that the sum of the numbers obtained is divisible by 7.
Let's list all possible outcomes of throwing two dice:
(1, 1), (1, 2), (1, 3), (1, 4), (1, 5), (1, 6)
(2, 1), (2, 2), (2, 3), (2, 4), (2, 5), (2, 6)
(3, 1), (3, 2), (3, 3), (3, 4), (3, 5), (3, 6)
(4, 1), (4, 2), (4, 3), (4, 4), (4, 5), (4, 6)
(5, 1), (5, 2), (5, 3), (5, 4), (5, 5), (5, 6)
(6, 1), (6, 2), (6, 3), (6, 4), (6, 5), (6, 6)
Now, let's find the pairs of outcomes whose sum is divisible by 7:
(1, 6), (2, 5), (3, 4), (4, 3), (5, 2), (6, 1)
There are 6 such pairs of outcomes.
Therefore, the required probability = 6/36 = 1/6.
Therefore, option 'A' is the correct answer.
When two dice are thrown simultaneously, the total number of outcomes = 6 x 6 = 36.
We need to find the probability that the sum of the numbers obtained is divisible by 7.
Let's list all possible outcomes of throwing two dice:
(1, 1), (1, 2), (1, 3), (1, 4), (1, 5), (1, 6)
(2, 1), (2, 2), (2, 3), (2, 4), (2, 5), (2, 6)
(3, 1), (3, 2), (3, 3), (3, 4), (3, 5), (3, 6)
(4, 1), (4, 2), (4, 3), (4, 4), (4, 5), (4, 6)
(5, 1), (5, 2), (5, 3), (5, 4), (5, 5), (5, 6)
(6, 1), (6, 2), (6, 3), (6, 4), (6, 5), (6, 6)
Now, let's find the pairs of outcomes whose sum is divisible by 7:
(1, 6), (2, 5), (3, 4), (4, 3), (5, 2), (6, 1)
There are 6 such pairs of outcomes.
Therefore, the required probability = 6/36 = 1/6.
Therefore, option 'A' is the correct answer.
Two linear and parallel RNA strands, defined by the equations 3x – 4y + 6 = 0 and 3x – 4y + 5 = 0 are hydrogen bonded together. The distance between the two strands is- a)0.2
- b)1.0
- c)1.2
- d)2
Correct answer is option 'A'. Can you explain this answer?
Two linear and parallel RNA strands, defined by the equations 3x – 4y + 6 = 0 and 3x – 4y + 5 = 0 are hydrogen bonded together. The distance between the two strands is
a)
0.2
b)
1.0
c)
1.2
d)
2

|
Raghav Rane answered |
Given:
Two linear and parallel RNA strands, defined by the equations 3x + 4y + 6 = 0 and 3x + 4y + 5 = 0 are hydrogen bonded together.
To find:
The distance between the two strands.
Solution:
The given equations represent two linear RNA strands. To find the distance between these strands, we need to find the perpendicular distance between them.
Step 1:
First, let's find the slope of the given lines by rearranging the equations in the slope-intercept form (y = mx + c).
Equation 1: 3x + 4y + 6 = 0
Rearranging, we get: 4y = -3x - 6
Dividing by 4, we get: y = -3/4x - 3/2
Equation 2: 3x + 4y + 5 = 0
Rearranging, we get: 4y = -3x - 5
Dividing by 4, we get: y = -3/4x - 5/4
Comparing the equations, we can see that the slopes of both lines are the same, i.e., -3/4. This indicates that the two RNA strands are parallel.
Step 2:
To find the distance between the parallel strands, we consider a point on one of the strands and find its perpendicular distance from the other strand.
Considering the point (0, -3/2) on the first strand (Equation 1), let's find its perpendicular distance from the second strand (Equation 2).
The perpendicular distance (d) can be found using the formula:
d = |ax + by + c| / √(a^2 + b^2)
Substituting the values from Equation 2, we get:
d = |3(0) + 4(-3/2) + 5| / √(3^2 + 4^2)
= |-6 + 5| / √(9 + 16)
= |-1| / √25
= 1 / 5
Therefore, the distance between the two RNA strands is 1 / 5, which is equal to 0.2.
Hence, the correct answer is option 'A' (0.2).
Two linear and parallel RNA strands, defined by the equations 3x + 4y + 6 = 0 and 3x + 4y + 5 = 0 are hydrogen bonded together.
To find:
The distance between the two strands.
Solution:
The given equations represent two linear RNA strands. To find the distance between these strands, we need to find the perpendicular distance between them.
Step 1:
First, let's find the slope of the given lines by rearranging the equations in the slope-intercept form (y = mx + c).
Equation 1: 3x + 4y + 6 = 0
Rearranging, we get: 4y = -3x - 6
Dividing by 4, we get: y = -3/4x - 3/2
Equation 2: 3x + 4y + 5 = 0
Rearranging, we get: 4y = -3x - 5
Dividing by 4, we get: y = -3/4x - 5/4
Comparing the equations, we can see that the slopes of both lines are the same, i.e., -3/4. This indicates that the two RNA strands are parallel.
Step 2:
To find the distance between the parallel strands, we consider a point on one of the strands and find its perpendicular distance from the other strand.
Considering the point (0, -3/2) on the first strand (Equation 1), let's find its perpendicular distance from the second strand (Equation 2).
The perpendicular distance (d) can be found using the formula:
d = |ax + by + c| / √(a^2 + b^2)
Substituting the values from Equation 2, we get:
d = |3(0) + 4(-3/2) + 5| / √(3^2 + 4^2)
= |-6 + 5| / √(9 + 16)
= |-1| / √25
= 1 / 5
Therefore, the distance between the two RNA strands is 1 / 5, which is equal to 0.2.
Hence, the correct answer is option 'A' (0.2).
If an aldol cleavage of glucose- 6- phosphate occurs in glycolysis, it will result in- a)products of equal carbon chain length
- b)products of unequal carbon chain length
- c)removal of phosphate group
- d)three C2 compounds
Correct answer is option 'B'. Can you explain this answer?
If an aldol cleavage of glucose- 6- phosphate occurs in glycolysis, it will result in
a)
products of equal carbon chain length
b)
products of unequal carbon chain length
c)
removal of phosphate group
d)
three C2 compounds

|
Vaibhav Ghosh answered |
Explanation:
In glycolysis, glucose-6-phosphate is converted into fructose-6-phosphate through a series of enzymatic reactions. However, under certain conditions, an aldol cleavage of glucose-6-phosphate can occur, resulting in the formation of two products.
Products of unequal carbon chain length:
The aldol cleavage of glucose-6-phosphate leads to the formation of two products: glyceraldehyde-3-phosphate (GAP) and dihydroxyacetone phosphate (DHAP). These two products have different carbon chain lengths.
Glyceraldehyde-3-phosphate (GAP):
GAP is a three-carbon compound. It can continue through the glycolytic pathway and participate in further reactions to generate ATP, NADH, and pyruvate.
Dihydroxyacetone phosphate (DHAP):
DHAP is a three-carbon compound as well, but it cannot directly continue through the glycolytic pathway. Instead, it needs to be converted into GAP by the enzyme triose phosphate isomerase. Once DHAP is converted into GAP, it can proceed through the glycolytic pathway.
Significance of aldol cleavage:
The aldol cleavage of glucose-6-phosphate is not a common occurrence in glycolysis. It usually happens under specific conditions, such as when the demand for NADPH is high. In such cases, DHAP can be converted into GAP through the action of the enzyme glycerol-3-phosphate dehydrogenase, generating NADPH. NADPH is an important coenzyme involved in various cellular processes, such as fatty acid synthesis and antioxidant defense.
Conclusion:
In summary, if an aldol cleavage of glucose-6-phosphate occurs in glycolysis, it will result in products of unequal carbon chain length, namely glyceraldehyde-3-phosphate (GAP) and dihydroxyacetone phosphate (DHAP). This cleavage provides an alternative pathway for the production of NADPH when it is needed in large quantities.
In glycolysis, glucose-6-phosphate is converted into fructose-6-phosphate through a series of enzymatic reactions. However, under certain conditions, an aldol cleavage of glucose-6-phosphate can occur, resulting in the formation of two products.
Products of unequal carbon chain length:
The aldol cleavage of glucose-6-phosphate leads to the formation of two products: glyceraldehyde-3-phosphate (GAP) and dihydroxyacetone phosphate (DHAP). These two products have different carbon chain lengths.
Glyceraldehyde-3-phosphate (GAP):
GAP is a three-carbon compound. It can continue through the glycolytic pathway and participate in further reactions to generate ATP, NADH, and pyruvate.
Dihydroxyacetone phosphate (DHAP):
DHAP is a three-carbon compound as well, but it cannot directly continue through the glycolytic pathway. Instead, it needs to be converted into GAP by the enzyme triose phosphate isomerase. Once DHAP is converted into GAP, it can proceed through the glycolytic pathway.
Significance of aldol cleavage:
The aldol cleavage of glucose-6-phosphate is not a common occurrence in glycolysis. It usually happens under specific conditions, such as when the demand for NADPH is high. In such cases, DHAP can be converted into GAP through the action of the enzyme glycerol-3-phosphate dehydrogenase, generating NADPH. NADPH is an important coenzyme involved in various cellular processes, such as fatty acid synthesis and antioxidant defense.
Conclusion:
In summary, if an aldol cleavage of glucose-6-phosphate occurs in glycolysis, it will result in products of unequal carbon chain length, namely glyceraldehyde-3-phosphate (GAP) and dihydroxyacetone phosphate (DHAP). This cleavage provides an alternative pathway for the production of NADPH when it is needed in large quantities.
Which one of the following statements is INCORRECT with respect to bacterial conjugation?- a)It facilitates transfer of genetic material
- b)It requires flagellum
- c)It can spread antibiotic resistance
- d)It can transfer virulence factors
Correct answer is option 'B'. Can you explain this answer?
Which one of the following statements is INCORRECT with respect to bacterial conjugation?
a)
It facilitates transfer of genetic material
b)
It requires flagellum
c)
It can spread antibiotic resistance
d)
It can transfer virulence factors

|
Niharika Kulkarni answered |
Bacterial Conjugation
Bacterial conjugation is a process of horizontal gene transfer between bacteria. It involves the transfer of genetic material from one bacterium to another through direct cell-to-cell contact. Here are the statements related to bacterial conjugation and their correctness:
Facilitates transfer of genetic material
Bacterial conjugation facilitates the transfer of genetic material, including plasmids and other genetic elements, from one bacterium to another. This transfer of genetic material can lead to the acquisition of new traits such as antibiotic resistance and virulence factors.
Requires flagellum
This statement is incorrect. Bacterial conjugation does not require a flagellum for transfer of genetic material. Instead, it involves the formation of a conjugation bridge or pilus, which is a specialized appendage that forms between the two bacterial cells. The pilus serves as a conduit for the transfer of genetic material from the donor to the recipient cell.
Can spread antibiotic resistance
Bacterial conjugation is one of the mechanisms by which antibiotic resistance genes can spread among bacteria. When a bacterium carrying antibiotic resistance genes conjugates with a bacterium that is susceptible to an antibiotic, the recipient cell can acquire the resistance genes and become resistant to the antibiotic as well.
Can transfer virulence factors
Bacterial conjugation can also transfer virulence factors, which are genes that enable bacteria to cause disease. The transfer of virulence factors can transform a non-pathogenic bacterium into a pathogenic one, leading to the development of infectious diseases.
Conclusion
Bacterial conjugation is an important mechanism of horizontal gene transfer in bacteria. It facilitates the transfer of genetic material, including antibiotic resistance genes and virulence factors, from one bacterium to another. It does not require a flagellum for transfer of genetic material.
Bacterial conjugation is a process of horizontal gene transfer between bacteria. It involves the transfer of genetic material from one bacterium to another through direct cell-to-cell contact. Here are the statements related to bacterial conjugation and their correctness:
Facilitates transfer of genetic material
Bacterial conjugation facilitates the transfer of genetic material, including plasmids and other genetic elements, from one bacterium to another. This transfer of genetic material can lead to the acquisition of new traits such as antibiotic resistance and virulence factors.
Requires flagellum
This statement is incorrect. Bacterial conjugation does not require a flagellum for transfer of genetic material. Instead, it involves the formation of a conjugation bridge or pilus, which is a specialized appendage that forms between the two bacterial cells. The pilus serves as a conduit for the transfer of genetic material from the donor to the recipient cell.
Can spread antibiotic resistance
Bacterial conjugation is one of the mechanisms by which antibiotic resistance genes can spread among bacteria. When a bacterium carrying antibiotic resistance genes conjugates with a bacterium that is susceptible to an antibiotic, the recipient cell can acquire the resistance genes and become resistant to the antibiotic as well.
Can transfer virulence factors
Bacterial conjugation can also transfer virulence factors, which are genes that enable bacteria to cause disease. The transfer of virulence factors can transform a non-pathogenic bacterium into a pathogenic one, leading to the development of infectious diseases.
Conclusion
Bacterial conjugation is an important mechanism of horizontal gene transfer in bacteria. It facilitates the transfer of genetic material, including antibiotic resistance genes and virulence factors, from one bacterium to another. It does not require a flagellum for transfer of genetic material.
Cyclic photophosphorylation produces ________- a)Energy only
- b)Reducing power only
- c)Both energy and reducing power
- d)Depending on the organism
Correct answer is option 'A'. Can you explain this answer?
Cyclic photophosphorylation produces ________
a)
Energy only
b)
Reducing power only
c)
Both energy and reducing power
d)
Depending on the organism

|
Seblewongel Girma answered |
In cyclic photophosphorylation (photo system I takes place) ,only ATP is formed.
A certain force applied to mass m1 given it an acceleration of 10 ms–2. The same force applied to mass m2 gives it an acceleration of 15ms–2. If the two masses are joined together and the same force is applied to the combination, the acceleration will be- a)6 ms–2
- b)3 ms–2
- c)9 ms–2
- d)12 ms–2
Correct answer is option 'A'. Can you explain this answer?
A certain force applied to mass m1 given it an acceleration of 10 ms–2. The same force applied to mass m2 gives it an acceleration of 15ms–2. If the two masses are joined together and the same force is applied to the combination, the acceleration will be
a)
6 ms–2
b)
3 ms–2
c)
9 ms–2
d)
12 ms–2
|
|
Arati Sathe answered |
12ms-2
The minimum light intensity that the human eye can perceive is 10-10 Wm-2. The area of the opening of our eye (the pupil) is approximately 4 cm-2. Consider yellow light with wavelength λ = 600 nm. The number of photons incident on the retina per second at the minimum intensity for the eye to respond is- a)1.5 × 103
- b)5 × 103
- c)8 × 103
- d)1.2 × 104
Correct answer is option 'D'. Can you explain this answer?
The minimum light intensity that the human eye can perceive is 10-10 Wm-2. The area of the opening of our eye (the pupil) is approximately 4 cm-2. Consider yellow light with wavelength λ = 600 nm. The number of photons incident on the retina per second at the minimum intensity for the eye to respond is
a)
1.5 × 103
b)
5 × 103
c)
8 × 103
d)
1.2 × 104

|
Rathin Das answered |

The population of a bacterial culture increases from one thousand to one billion in five hours. The doubling time of the culture (correct to 1 decimal place) is _______ min.
Correct answer is between '14.0,16.0'. Can you explain this answer?
The population of a bacterial culture increases from one thousand to one billion in five hours. The doubling time of the culture (correct to 1 decimal place) is _______ min.

|
Bhavana Dasgupta answered |
Explanation:
The formula for calculating the doubling time (td) is given by:
td = ln(2) / r
Where, ln(2) is the natural logarithm of 2 and r is the growth rate.
Step 1: Calculate the growth rate
The growth rate (r) can be calculated using the formula:
r = (ln(Nt) - ln(N0)) / t
Where, Nt is the final population, N0 is the initial population and t is the time taken for the population to grow.
Given, Nt = 1 billion, N0 = 1000 and t = 5 hours
r = (ln(1 billion) - ln(1000)) / 5
r = 29.1 per hour
Step 2: Calculate the doubling time
td = ln(2) / r
td = ln(2) / 29.1
td = 0.0238 hours (convert to minutes)
td = 0.0238 x 60 = 1.43 minutes
Therefore, the doubling time of the bacterial culture is 1.43 minutes. Rounded to one decimal place, the answer is between 14.0 and 16.0 minutes.
The formula for calculating the doubling time (td) is given by:
td = ln(2) / r
Where, ln(2) is the natural logarithm of 2 and r is the growth rate.
Step 1: Calculate the growth rate
The growth rate (r) can be calculated using the formula:
r = (ln(Nt) - ln(N0)) / t
Where, Nt is the final population, N0 is the initial population and t is the time taken for the population to grow.
Given, Nt = 1 billion, N0 = 1000 and t = 5 hours
r = (ln(1 billion) - ln(1000)) / 5
r = 29.1 per hour
Step 2: Calculate the doubling time
td = ln(2) / r
td = ln(2) / 29.1
td = 0.0238 hours (convert to minutes)
td = 0.0238 x 60 = 1.43 minutes
Therefore, the doubling time of the bacterial culture is 1.43 minutes. Rounded to one decimal place, the answer is between 14.0 and 16.0 minutes.
The pH of a 0.1 M solution of monosodium succinate (pKa1 = 4.19 and pKa2 = 5.57) is ____
Correct answer is '4.8'. Can you explain this answer?
The pH of a 0.1 M solution of monosodium succinate (pKa1 = 4.19 and pKa2 = 5.57) is ____

|
Soumya Sharma answered |
Explanation:
Monosodium succinate is a weak acid with two acidic protons. The given pKa values are pKa1 = 4.19 and pKa2 = 5.57. The dissociation reactions are as follows:
HSucc ⇌ H+ + Succ- Ka1 = 10^-4.19
Succ- ⇌ H+ + Succ2- Ka2 = 10^-5.57
Step 1: Calculate the pH at half-equivalence point:
The half-equivalence point is when [H+] = Ka1. At this point, half of the monosodium succinate has been converted to its conjugate base, and the other half remains in its acidic form.
Ka1 = [H+][Succ-]/[HSucc]
10^-4.19 = [H+][0.1]/[0.1]
[H+] = 10^-4.19
pH = 4.19
Step 2: Calculate the pH at the equivalence point:
The equivalence point is when all the monosodium succinate has been converted to its conjugate base, Succ2-. At this point, [Succ-] = [Succ2-], and the solution contains only the conjugate base.
Ka2 = [H+][Succ2-]/[Succ-]
10^-5.57 = [H+][0.1]/[0]
[H+] = infinity (pH = -log[H+] = infinity)
The pH at the equivalence point is greater than 7, indicating a basic solution.
Step 3: Calculate the pH at the midway between the two equivalence points:
The midway between the two equivalence points is when [Succ2-] = Ka1 and [HSucc] = Ka2. At this point, the solution contains equal amounts of the conjugate base and the acidic form.
Ka1 = [H+][Succ-]/[HSucc]
10^-4.19 = [H+][0.05]/[0.05]
[H+] = 10^-4.19
Ka2 = [H+][Succ2-]/[Succ-]
10^-5.57 = [H+][0.05]/[0.05]
[H+] = 10^-5.57
The average of the two [H+] values is:
[H+]avg = (10^-4.19 + 10^-5.57)/2 = 2.83 x 10^-5
pH = -log[H+]avg = 4.8
Therefore, the pH of a 0.1 M solution of monosodium succinate is 4.8.
In metal- carbonyl complexes, the p- back bonding is- a)pπ – dπ type
- b)dπ – dπ type
- c)dπ – π* type
- d)dπ – σ* type
Correct answer is option 'C'. Can you explain this answer?
In metal- carbonyl complexes, the p- back bonding is
a)
pπ – dπ type
b)
dπ – dπ type
c)
dπ – π* type
d)
dπ – σ* type

|
Rutuja Choudhary answered |
Understanding p-Back Bonding in Metal-Carbonyl Complexes
Metal-carbonyl complexes are significant in coordination chemistry, primarily due to their unique electronic properties. One important aspect of these properties is the phenomenon known as p-back bonding.
What is p-Back Bonding?
- P-back bonding refers to the electron donation from filled metal d-orbitals to empty p-orbitals of the CO ligand.
- This process enhances the stability of the metal-carbonyl complex and helps in lowering the CO stretching frequency.
Role of d * Orbitals
- In metal-carbonyl complexes, the metal center typically has d-electrons that can participate in back donation.
- The d * orbitals, which are the higher-energy d-orbitals, are crucial for this bonding.
Why d * Type?
- The d * orbitals have the appropriate symmetry and energy to overlap effectively with the π* (pi-star) orbitals of carbon monoxide.
- This overlap allows for effective electron transfer from the metal to the ligand, stabilizing the overall complex.
Conclusion
- Therefore, in metal-carbonyl complexes, the p-back bonding mechanism primarily involves d * type orbitals.
- This contributes significantly to the properties and reactivity of these complexes, making option 'C' the correct choice.
Understanding these concepts is vital for comprehending how metal-carbonyl complexes function in various chemical processes and applications.
Metal-carbonyl complexes are significant in coordination chemistry, primarily due to their unique electronic properties. One important aspect of these properties is the phenomenon known as p-back bonding.
What is p-Back Bonding?
- P-back bonding refers to the electron donation from filled metal d-orbitals to empty p-orbitals of the CO ligand.
- This process enhances the stability of the metal-carbonyl complex and helps in lowering the CO stretching frequency.
Role of d * Orbitals
- In metal-carbonyl complexes, the metal center typically has d-electrons that can participate in back donation.
- The d * orbitals, which are the higher-energy d-orbitals, are crucial for this bonding.
Why d * Type?
- The d * orbitals have the appropriate symmetry and energy to overlap effectively with the π* (pi-star) orbitals of carbon monoxide.
- This overlap allows for effective electron transfer from the metal to the ligand, stabilizing the overall complex.
Conclusion
- Therefore, in metal-carbonyl complexes, the p-back bonding mechanism primarily involves d * type orbitals.
- This contributes significantly to the properties and reactivity of these complexes, making option 'C' the correct choice.
Understanding these concepts is vital for comprehending how metal-carbonyl complexes function in various chemical processes and applications.
Pick the correct statement(s) with respect to the inter-conversion of the topoisomers of a circularlyclosed double stranded DNA.- a)Only one strand needs to be cut
- b)Both strands have to be cut
- c)No strand needs to be cut
- d)ATP is required for inter-conversion
Correct answer is option 'A,D'. Can you explain this answer?
Pick the correct statement(s) with respect to the inter-conversion of the topoisomers of a circularlyclosed double stranded DNA.
a)
Only one strand needs to be cut
b)
Both strands have to be cut
c)
No strand needs to be cut
d)
ATP is required for inter-conversion

|
Kiran Pillai answered |
Correct Answer :- a,d
Explanation : Answer is A & D. only one strand needs to be cut. As it will bring DNA from Positively supercoiled to negative supercoiled. ATP is required for inter-conversion. For Positive to negative supercoiled conditions.
The refractive index of diamond is 2.419. If the speed of light in vacuum is 3 × 108 m s–1, then the speed of light in diamond is- a)1.240 × 108 m s–1
- b)1.352 × 108 m s–1
- c)1.521 × 108 m s–1
- d)2.433 × 108 m s–1
Correct answer is option 'A'. Can you explain this answer?
The refractive index of diamond is 2.419. If the speed of light in vacuum is 3 × 108 m s–1, then the speed of light in diamond is
a)
1.240 × 108 m s–1
b)
1.352 × 108 m s–1
c)
1.521 × 108 m s–1
d)
2.433 × 108 m s–1

|
Shruti Datta answered |
Calculation of the speed of light in diamond
Refractive index of diamond = 2.419
Speed of light in vacuum = 3 x 10^8 m/s
The formula relating the speed of light in a medium to the speed of light in vacuum and the refractive index of the medium is:
speed of light in medium = speed of light in vacuum / refractive index of the medium
Substituting the given values into the formula, we get:
speed of light in diamond = (3 x 10^8 m/s) / 2.419
= 1.240 x 10^8 m/s
Therefore, the speed of light in diamond is 1.240 x 10^8 m/s, which is option A.
Explanation
The refractive index of a medium is a measure of how much the speed of light is reduced when it passes through the medium. The higher the refractive index, the more the speed of light is reduced. Diamond has a very high refractive index of 2.419, which means that the speed of light in diamond is much lower than the speed of light in vacuum.
The formula relating the speed of light in a medium to the speed of light in vacuum and the refractive index of the medium is derived from Snell's law of refraction. When light passes from one medium to another, it changes direction due to the change in speed. Snell's law relates the angle of incidence and the angle of refraction to the refractive indices of the two media.
In this question, we are given the refractive index of diamond and the speed of light in vacuum. Using the formula, we can calculate the speed of light in diamond. The answer is option A, 1.240 x 10^8 m/s.
Refractive index of diamond = 2.419
Speed of light in vacuum = 3 x 10^8 m/s
The formula relating the speed of light in a medium to the speed of light in vacuum and the refractive index of the medium is:
speed of light in medium = speed of light in vacuum / refractive index of the medium
Substituting the given values into the formula, we get:
speed of light in diamond = (3 x 10^8 m/s) / 2.419
= 1.240 x 10^8 m/s
Therefore, the speed of light in diamond is 1.240 x 10^8 m/s, which is option A.
Explanation
The refractive index of a medium is a measure of how much the speed of light is reduced when it passes through the medium. The higher the refractive index, the more the speed of light is reduced. Diamond has a very high refractive index of 2.419, which means that the speed of light in diamond is much lower than the speed of light in vacuum.
The formula relating the speed of light in a medium to the speed of light in vacuum and the refractive index of the medium is derived from Snell's law of refraction. When light passes from one medium to another, it changes direction due to the change in speed. Snell's law relates the angle of incidence and the angle of refraction to the refractive indices of the two media.
In this question, we are given the refractive index of diamond and the speed of light in vacuum. Using the formula, we can calculate the speed of light in diamond. The answer is option A, 1.240 x 10^8 m/s.
The technique that involves impacting samples with electrons is _______.- a)NMR spectroscopy
- b)ESI mass spectrometry
- c)IR spectroscopy
- d)UV-vis spectroscopy
Correct answer is option 'B'. Can you explain this answer?
The technique that involves impacting samples with electrons is _______.
a)
NMR spectroscopy
b)
ESI mass spectrometry
c)
IR spectroscopy
d)
UV-vis spectroscopy

|
Varun Yadav answered |
ESI mass spectrometry
ESI stands for Electrospray Ionization, which is a technique used in mass spectrometry. Mass spectrometry is an analytical technique used to determine the molecular weight and structure of a sample. It involves the ionization of molecules and the separation of these ions based on their mass-to-charge ratio.
Principle of ESI Mass Spectrometry:
Electrospray ionization involves the generation of charged droplets from a liquid sample. These droplets are then subjected to a strong electric field, causing them to break apart into smaller droplets. During this process, the solvent molecules evaporate, and the remaining charged analyte molecules are transferred into the gas phase as ions.
The Process:
1. Sample preparation: The sample is dissolved in a suitable solvent and introduced into the mass spectrometer through a capillary.
2. Electrospray: The sample solution is passed through a needle connected to a high voltage power supply. The high voltage creates a strong electric field that causes the formation of a fine aerosol of charged droplets.
3. Desolvation: As the droplets travel through the mass spectrometer, the solvent molecules evaporate, leaving behind the analyte ions.
4. Ionization: The analyte molecules acquire a charge by either gaining or losing electrons. This can be achieved by introducing a reagent gas, such as ammonia or acetonitrile, into the mass spectrometer.
5. Ion separation: The ions are then guided into the mass analyzer, where they are separated based on their mass-to-charge ratio. This separation can be achieved using various techniques, such as time-of-flight (TOF), quadrupole, or ion trap.
6. Ion detection: The separated ions are detected by a detector, which generates an electrical signal proportional to the abundance of each ion.
7. Data analysis: The recorded data is analyzed to determine the mass-to-charge ratio of the ions and their relative abundance. This information can be used to identify the molecular weight and structure of the sample.
Advantages of ESI Mass Spectrometry:
- ESI is compatible with a wide range of analytes, including small organic molecules, peptides, proteins, and nucleic acids.
- It can provide information about the molecular weight, structure, and fragmentation patterns of the analyte.
- ESI is a soft ionization technique, meaning it produces minimal fragmentation of the analyte ions, allowing for the detection of intact molecular ions.
- It is highly sensitive, capable of detecting analytes in the picogram to femtogram range.
In conclusion, ESI mass spectrometry is a technique that involves impacting samples with electrons to generate ions for analysis. It is widely used in various fields, including chemistry, biology, and medicine, for the identification and characterization of molecules.
ESI stands for Electrospray Ionization, which is a technique used in mass spectrometry. Mass spectrometry is an analytical technique used to determine the molecular weight and structure of a sample. It involves the ionization of molecules and the separation of these ions based on their mass-to-charge ratio.
Principle of ESI Mass Spectrometry:
Electrospray ionization involves the generation of charged droplets from a liquid sample. These droplets are then subjected to a strong electric field, causing them to break apart into smaller droplets. During this process, the solvent molecules evaporate, and the remaining charged analyte molecules are transferred into the gas phase as ions.
The Process:
1. Sample preparation: The sample is dissolved in a suitable solvent and introduced into the mass spectrometer through a capillary.
2. Electrospray: The sample solution is passed through a needle connected to a high voltage power supply. The high voltage creates a strong electric field that causes the formation of a fine aerosol of charged droplets.
3. Desolvation: As the droplets travel through the mass spectrometer, the solvent molecules evaporate, leaving behind the analyte ions.
4. Ionization: The analyte molecules acquire a charge by either gaining or losing electrons. This can be achieved by introducing a reagent gas, such as ammonia or acetonitrile, into the mass spectrometer.
5. Ion separation: The ions are then guided into the mass analyzer, where they are separated based on their mass-to-charge ratio. This separation can be achieved using various techniques, such as time-of-flight (TOF), quadrupole, or ion trap.
6. Ion detection: The separated ions are detected by a detector, which generates an electrical signal proportional to the abundance of each ion.
7. Data analysis: The recorded data is analyzed to determine the mass-to-charge ratio of the ions and their relative abundance. This information can be used to identify the molecular weight and structure of the sample.
Advantages of ESI Mass Spectrometry:
- ESI is compatible with a wide range of analytes, including small organic molecules, peptides, proteins, and nucleic acids.
- It can provide information about the molecular weight, structure, and fragmentation patterns of the analyte.
- ESI is a soft ionization technique, meaning it produces minimal fragmentation of the analyte ions, allowing for the detection of intact molecular ions.
- It is highly sensitive, capable of detecting analytes in the picogram to femtogram range.
In conclusion, ESI mass spectrometry is a technique that involves impacting samples with electrons to generate ions for analysis. It is widely used in various fields, including chemistry, biology, and medicine, for the identification and characterization of molecules.
In a class of 100 students there are 70 boys whose average marks in a subject are 75. If the average marks of the complete class is 72, then what is the average marks of the girls ?
Correct answer is '65'. Can you explain this answer?
In a class of 100 students there are 70 boys whose average marks in a subject are 75. If the average marks of the complete class is 72, then what is the average marks of the girls ?

|
Baishali Bajaj answered |
Number of girls = 100 - 70 = 30
Total marks of 100 students = 100 x 72 = 7200
Total marks of 70 boys = 70 x 75 = 5250
This implies that total marks of 30 girls = 7200 - 5250 = 1950
Therefore, average marks of girls = 1950/30 = 65
An example of a eukaryotic chemoorganotroph microorganism lacking chlorophyll and having mycelial thallus is- a)yeast
- b)bacteria
- c)fungi
- d)protozoa
Correct answer is option 'C'. Can you explain this answer?
An example of a eukaryotic chemoorganotroph microorganism lacking chlorophyll and having mycelial thallus is
a)
yeast
b)
bacteria
c)
fungi
d)
protozoa
|
|
Sarita Yadav answered |
Archaea and bacteria are classified as prokaryotes because they lack a cellular nucleus. Protozoa are unicellular organisms with complex cell structures; most are motile. Microscopic fungi include molds and yeasts. Helminths are multicellular parasitic worms.
How many ATP equivalents per mole of glucose input are required for gluconeogenesis ?- a)3
- b)6
- c)10
- d)4
Correct answer is option 'B'. Can you explain this answer?
How many ATP equivalents per mole of glucose input are required for gluconeogenesis ?
a)
3
b)
6
c)
10
d)
4

|
Madhavan Iyer answered |
Ans.
Option (b)
You know how glycolysis uses a total number of 2 atp and produces (2+2) 4 atps. So we know that gluconegenesis is the reverse of glycolysis and we know that the last step which produced 2 atp by converting pep to pyruvate is now divided into two steps of their own (pyruvate-> oaa and oaa-> pep).
Glycolysis is like this : 2 + 2 = 4.
Gluconeogenesis : 2 + (2 atp +2 gtp) = 6 which means we need a total of 6 to go ahead.
Amongst the following, the elongated, fibrous protein is- a)Myoglobin
- b)Keratin
- c)Albumin
- d)Calmodulim
Correct answer is option 'B'. Can you explain this answer?
Amongst the following, the elongated, fibrous protein is
a)
Myoglobin
b)
Keratin
c)
Albumin
d)
Calmodulim

|
Anirban Khanna answered |
Fibrous proteins, also called scleroproteins, are long filamentous protein molecules.[1]
Fibrous proteins are only found in animals.
Fibrous proteins form 'rod' or 'wire' -like shapes and are usually inert structural or storage proteins. They are generally water-insoluble. Fibrous proteins are usually used to construct connective tissues, tendons, bone and muscle fiber.
Examples of fibrous proteins include keratins, collagens and elastins.
Fingernails and claws are made up of the common fibrous proteins, Keratin.
The dimensions of coefficient of viscosity are _______.- a)ML-1T-1
- b)ML-1T-2
- c)ML-2T-2
- d)ML-2T-1
Correct answer is option 'A'. Can you explain this answer?
The dimensions of coefficient of viscosity are _______.
a)
ML-1T-1
b)
ML-1T-2
c)
ML-2T-2
d)
ML-2T-1

|
Kanchan Butola answered |
Coefficient of viscosity, η=Fdx/Adv
force F=[MLT^-2]
Area A=[L^2]
dv/dx=[LT^-1]/L=T^-1
Coefficient of viscosity, η=[MLT^-2]/[L^2][T^-1]
Coefficient of viscosity, η=[ML^-1T^-1]
force F=[MLT^-2]
Area A=[L^2]
dv/dx=[LT^-1]/L=T^-1
Coefficient of viscosity, η=[MLT^-2]/[L^2][T^-1]
Coefficient of viscosity, η=[ML^-1T^-1]
A particle starting from rest is subjected to a constant force. The plot of distance traveled along thedirection of the force as a function of time is a/an ______.- a)straight line
- b)circle
- c)parabola
- d)ellipse
Correct answer is option 'C'. Can you explain this answer?
A particle starting from rest is subjected to a constant force. The plot of distance traveled along thedirection of the force as a function of time is a/an ______.
a)
straight line
b)
circle
c)
parabola
d)
ellipse

|
Rutuja Sengupta answered |
**Explanation:**
When a particle is subjected to a constant force, its distance traveled along the direction of the force as a function of time can be represented by a parabolic plot.
**1. Motion under Constant Force:**
When a particle is subjected to a constant force, it undergoes accelerated motion. The acceleration of the particle is given by Newton's second law of motion:
F = ma
where F is the force acting on the particle, m is the mass of the particle, and a is the acceleration of the particle.
**2. Distance as a Function of Time:**
The distance traveled by the particle along the direction of the force can be determined by integrating the velocity of the particle with respect to time:
s = ∫v dt
where s is the distance traveled, v is the velocity of the particle, and t is the time.
**3. Relationship between Velocity and Time:**
The velocity of the particle can be determined by integrating the acceleration of the particle with respect to time:
v = ∫a dt
Substituting the value of acceleration from Newton's second law of motion, we get:
v = ∫(F/m) dt
**4. Relationship between Distance and Time:**
Substituting the value of velocity in the equation for distance, we get:
s = ∫(∫(F/m) dt) dt
Simplifying the equation, we get:
s = (1/2)(F/m)t^2
This equation represents a parabolic plot of distance traveled along the direction of the force as a function of time. Hence, the correct answer is option 'C' - parabola.
When a particle is subjected to a constant force, its distance traveled along the direction of the force as a function of time can be represented by a parabolic plot.
**1. Motion under Constant Force:**
When a particle is subjected to a constant force, it undergoes accelerated motion. The acceleration of the particle is given by Newton's second law of motion:
F = ma
where F is the force acting on the particle, m is the mass of the particle, and a is the acceleration of the particle.
**2. Distance as a Function of Time:**
The distance traveled by the particle along the direction of the force can be determined by integrating the velocity of the particle with respect to time:
s = ∫v dt
where s is the distance traveled, v is the velocity of the particle, and t is the time.
**3. Relationship between Velocity and Time:**
The velocity of the particle can be determined by integrating the acceleration of the particle with respect to time:
v = ∫a dt
Substituting the value of acceleration from Newton's second law of motion, we get:
v = ∫(F/m) dt
**4. Relationship between Distance and Time:**
Substituting the value of velocity in the equation for distance, we get:
s = ∫(∫(F/m) dt) dt
Simplifying the equation, we get:
s = (1/2)(F/m)t^2
This equation represents a parabolic plot of distance traveled along the direction of the force as a function of time. Hence, the correct answer is option 'C' - parabola.
A mutation in the operator locus of lac operon that confers constitutive expression of β-galactosidase is ________.- a)cis dominant
- b)trans dominant
- c)co-dominant
- d)dominant negative
Correct answer is option 'A'. Can you explain this answer?
A mutation in the operator locus of lac operon that confers constitutive expression of β-galactosidase is ________.
a)
cis dominant
b)
trans dominant
c)
co-dominant
d)
dominant negative
|
|
Chirag Verma answered |
A mutation in the operator locus of lac operon that confers constitutive expression of β-galactosidase is
cis dominant
Which of the following is NOT involved in eukaryotic translation ?- a)Ribosome
- b)Spliceosome
- c)mRNA
- d)tRNA
Correct answer is option 'B'. Can you explain this answer?
Which of the following is NOT involved in eukaryotic translation ?
a)
Ribosome
b)
Spliceosome
c)
mRNA
d)
tRNA

|
Mahi Dasgupta answered |
Eukaryotic Translation and Spliceosome
Eukaryotic translation is the process by which proteins are synthesized from mRNA templates. It involves several components, including ribosomes, mRNA, tRNA, and various protein factors. However, spliceosome is not involved in eukaryotic translation.
Ribosome
Ribosomes are the cellular machines that carry out protein synthesis. They consist of two subunits, each of which contains rRNA and multiple ribosomal proteins. The small subunit binds to mRNA, while the large subunit catalyzes peptide bond formation between amino acids.
mRNA
mRNA is the intermediate molecule that carries the genetic information from DNA to ribosomes. It is produced by transcription, during which a portion of DNA is copied into RNA by RNA polymerase.
tRNA
tRNA is a small RNA molecule that serves as an adapter between mRNA and amino acids. It contains an anticodon that pairs with a complementary codon on mRNA, as well as an amino acid attachment site that binds to a specific amino acid.
Spliceosome
Spliceosome is a complex of RNA and protein molecules that removes introns from pre-mRNA and joins exons together to form mature mRNA. This process is called splicing and it occurs in the nucleus before mRNA is exported to the cytoplasm for translation.
Conclusion
In summary, eukaryotic translation involves ribosomes, mRNA, and tRNA, but not spliceosome. Ribosomes are responsible for peptide bond formation between amino acids, mRNA carries genetic information from DNA to ribosomes, and tRNA serves as an adapter between mRNA and amino acids. Spliceosome, on the other hand, is involved in pre-mRNA processing, not translation.
Eukaryotic translation is the process by which proteins are synthesized from mRNA templates. It involves several components, including ribosomes, mRNA, tRNA, and various protein factors. However, spliceosome is not involved in eukaryotic translation.
Ribosome
Ribosomes are the cellular machines that carry out protein synthesis. They consist of two subunits, each of which contains rRNA and multiple ribosomal proteins. The small subunit binds to mRNA, while the large subunit catalyzes peptide bond formation between amino acids.
mRNA
mRNA is the intermediate molecule that carries the genetic information from DNA to ribosomes. It is produced by transcription, during which a portion of DNA is copied into RNA by RNA polymerase.
tRNA
tRNA is a small RNA molecule that serves as an adapter between mRNA and amino acids. It contains an anticodon that pairs with a complementary codon on mRNA, as well as an amino acid attachment site that binds to a specific amino acid.
Spliceosome
Spliceosome is a complex of RNA and protein molecules that removes introns from pre-mRNA and joins exons together to form mature mRNA. This process is called splicing and it occurs in the nucleus before mRNA is exported to the cytoplasm for translation.
Conclusion
In summary, eukaryotic translation involves ribosomes, mRNA, and tRNA, but not spliceosome. Ribosomes are responsible for peptide bond formation between amino acids, mRNA carries genetic information from DNA to ribosomes, and tRNA serves as an adapter between mRNA and amino acids. Spliceosome, on the other hand, is involved in pre-mRNA processing, not translation.
Which one of the following remains unchanged when light waves enter water from air?- a)Wavelength
- b)Wavenumber
- c)Frequency
- d)Intensity
Correct answer is option 'C'. Can you explain this answer?
Which one of the following remains unchanged when light waves enter water from air?
a)
Wavelength
b)
Wavenumber
c)
Frequency
d)
Intensity
|
|
Vikram Kapoor answered |
Frequency of a wave is characterised by the source of wave, therefore it will not be affected by change in medium .
In a large wild flower population, assume that no new mutations occur and that no natural selection operates. What factor(s) will affect the frequency of a genotype in this population ?- a)Non- random mating
- b)Gene flow
- c)Out- breeding within the population
- d)Invasion of a new pathogen that kills a large number of individuals in the population
Correct answer is option 'A,B,C,D'. Can you explain this answer?
In a large wild flower population, assume that no new mutations occur and that no natural selection operates. What factor(s) will affect the frequency of a genotype in this population ?
a)
Non- random mating
b)
Gene flow
c)
Out- breeding within the population
d)
Invasion of a new pathogen that kills a large number of individuals in the population

|
Bhavana Pillai answered |
Factors affecting genotype frequency in a large wildflower population
Non-random mating:
- Non-random mating can occur when individuals choose their mates based on specific traits, such as color or size.
- This can lead to an increase in the frequency of certain genotypes if individuals with those genotypes are preferred as mates.
- Over time, this can result in a decrease in genetic diversity within the population.
Gene flow:
- Gene flow occurs when individuals move between populations and bring their genes with them.
- This can introduce new genotypes into the population and increase genetic diversity.
- Conversely, it can also lead to a decrease in the frequency of certain genotypes if individuals with those genotypes leave the population.
Outbreeding within the population:
- Outbreeding occurs when individuals mate with individuals from other populations or with unrelated individuals within their own population.
- This can increase genetic diversity within the population and lead to a decrease in the frequency of certain genotypes.
Invasion of a new pathogen:
- The invasion of a new pathogen that kills a large number of individuals in the population can lead to a decrease in the frequency of certain genotypes.
- If individuals with those genotypes are more susceptible to the pathogen, their frequency in the population will decrease.
- This can result in a decrease in genetic diversity within the population.
Conclusion:
In a large wildflower population, various factors can affect the frequency of a genotype. Non-random mating, gene flow, outbreeding within the population, and invasion of a new pathogen are all factors that can impact the frequency of a genotype in the population. Understanding these factors is important for understanding the genetic diversity and evolution of populations.
Non-random mating:
- Non-random mating can occur when individuals choose their mates based on specific traits, such as color or size.
- This can lead to an increase in the frequency of certain genotypes if individuals with those genotypes are preferred as mates.
- Over time, this can result in a decrease in genetic diversity within the population.
Gene flow:
- Gene flow occurs when individuals move between populations and bring their genes with them.
- This can introduce new genotypes into the population and increase genetic diversity.
- Conversely, it can also lead to a decrease in the frequency of certain genotypes if individuals with those genotypes leave the population.
Outbreeding within the population:
- Outbreeding occurs when individuals mate with individuals from other populations or with unrelated individuals within their own population.
- This can increase genetic diversity within the population and lead to a decrease in the frequency of certain genotypes.
Invasion of a new pathogen:
- The invasion of a new pathogen that kills a large number of individuals in the population can lead to a decrease in the frequency of certain genotypes.
- If individuals with those genotypes are more susceptible to the pathogen, their frequency in the population will decrease.
- This can result in a decrease in genetic diversity within the population.
Conclusion:
In a large wildflower population, various factors can affect the frequency of a genotype. Non-random mating, gene flow, outbreeding within the population, and invasion of a new pathogen are all factors that can impact the frequency of a genotype in the population. Understanding these factors is important for understanding the genetic diversity and evolution of populations.
The standard oxidation potentials for oxidation of NADH and H2O are + 0.315 V and – 0.815 V, respectively. The standard free energy for oxidation of 1 mole of NADH by oxygen under standard conditions (correct to 1 decimal place) is _______ kJ. [Faraday Constant is 96500 C mole–1]
Correct answer is between '-219.0,-217.0'. Can you explain this answer?
The standard oxidation potentials for oxidation of NADH and H2O are + 0.315 V and – 0.815 V, respectively. The standard free energy for oxidation of 1 mole of NADH by oxygen under standard conditions (correct to 1 decimal place) is _______ kJ. [Faraday Constant is 96500 C mole–1]

|
Sneha Menon answered |
Standard Free Energy for Oxidation of NADH by Oxygen
Introduction: The standard free energy change (∆G°) for a chemical reaction is a measure of the maximum amount of work that can be obtained from the reaction at constant temperature and pressure.
Given:
- Standard oxidation potential for NADH (E°NADH) = 0.315 V
- Standard oxidation potential for H2O (E°H2O) = 0.815 V
- Faraday constant (F) = 96500 C mole^-1
Formula: The standard free energy change (∆G°) for a chemical reaction can be calculated using the following formula:
∆G° = -nFE°
Where,
n = number of electrons transferred in the reaction
F = Faraday constant
E° = standard oxidation potential
Calculation:
The balanced equation for the oxidation of NADH by oxygen is:
NADH + ½O2 + H+ → NAD+ + H2O
The number of electrons transferred in the reaction is 2 (from NADH to O2). Therefore, n = 2.
The standard free energy change for the reaction can be calculated as follows:
∆G° = -nFE°
∆G° = -2 x F x (E°H2O - E°NADH)
∆G° = -2 x 96500 x (0.815 - 0.315)
∆G° = -2 x 96500 x 0.5
∆G° = -96,500 J mole^-1
To convert J mole^-1 to kJ mole^-1, we divide by 1000:
∆G° = -96.5 kJ mole^-1
Rounding off to one decimal place, we get:
∆G° = -96.5 kJ mole^-1 ≈ -219.0 kJ mole^-1
Therefore, the standard free energy for oxidation of 1 mole of NADH by oxygen under standard conditions is approximately -219.0 kJ.
Introduction: The standard free energy change (∆G°) for a chemical reaction is a measure of the maximum amount of work that can be obtained from the reaction at constant temperature and pressure.
Given:
- Standard oxidation potential for NADH (E°NADH) = 0.315 V
- Standard oxidation potential for H2O (E°H2O) = 0.815 V
- Faraday constant (F) = 96500 C mole^-1
Formula: The standard free energy change (∆G°) for a chemical reaction can be calculated using the following formula:
∆G° = -nFE°
Where,
n = number of electrons transferred in the reaction
F = Faraday constant
E° = standard oxidation potential
Calculation:
The balanced equation for the oxidation of NADH by oxygen is:
NADH + ½O2 + H+ → NAD+ + H2O
The number of electrons transferred in the reaction is 2 (from NADH to O2). Therefore, n = 2.
The standard free energy change for the reaction can be calculated as follows:
∆G° = -nFE°
∆G° = -2 x F x (E°H2O - E°NADH)
∆G° = -2 x 96500 x (0.815 - 0.315)
∆G° = -2 x 96500 x 0.5
∆G° = -96,500 J mole^-1
To convert J mole^-1 to kJ mole^-1, we divide by 1000:
∆G° = -96.5 kJ mole^-1
Rounding off to one decimal place, we get:
∆G° = -96.5 kJ mole^-1 ≈ -219.0 kJ mole^-1
Therefore, the standard free energy for oxidation of 1 mole of NADH by oxygen under standard conditions is approximately -219.0 kJ.
Let U= {1, 2, 3, 4, 5.} A subset S is chosen uniformly at random from the non-empty subsets of U. What is the probability that S does NOT have two consecutive elements?- a)9/31
- b)10/31
- c)11/31
- d)12/31
Correct answer is option 'D'. Can you explain this answer?
Let U= {1, 2, 3, 4, 5.} A subset S is chosen uniformly at random from the non-empty subsets of U. What is the probability that S does NOT have two consecutive elements?
a)
9/31
b)
10/31
c)
11/31
d)
12/31
|
|
Chirag Verma answered |
Given
u={1,2,3,4,5}.
a non-empty subset of U. = 31
1 , 2 , 3 , 4 , 5 , (1,2) ,(1,3) ,(1,4) ,(1,5) ,(2,3) ,(2,4) (2,5) ,(3,4),(3,5) ,(4,5) ,(1,2 ,3) ,(1,2,4) ,(1,2,5) , (1,3 ,4) ,(1,3,5) ,( 1,4,5) ,(2,3 ,4) ,(2,3,5) ,(2,4,5) ,(3,4,5) ,(1,2,3,4) ,(1,2,3,5) (1,2,4,5),(1,3,4,5) ,(2,3,4,5) ,(1,2,3,4,5)
A subset S is chosen uniformly at random from a does not have two consecutive elements
= 12
probability at S does not have two consecutive elements = (12/31)
Proinsulin is an 84 residue polypeptide with six cysteines. How many different disulfide combinations are possible?
Correct answer is '15 to 15'. Can you explain this answer?
Proinsulin is an 84 residue polypeptide with six cysteines. How many different disulfide combinations are possible?

|
Akanksha Choudhary answered |
Possible Disulfide Combinations in Proinsulin
Introduction:
Proinsulin is an 84 residue polypeptide with six cysteines. The disulfide bonds formed between the cysteine residues are critical for the correct folding and function of the protein. The number of possible disulfide combinations in proinsulin can be calculated using a combinatorial approach.
Method:
The number of possible disulfide combinations can be calculated using the formula:
nCr = n! / r! (n-r)!
Where n is the total number of cysteine residues and r is the number of disulfide bonds formed.
Calculation:
In proinsulin, there are six cysteine residues. To determine the number of possible disulfide combinations, we need to consider the number of disulfide bonds that can be formed.
- Number of disulfide bonds = 0: There is only one possible combination where all cysteine residues remain unpaired.
- Number of disulfide bonds = 1: There are six possible combinations where one cysteine residue pairs with another cysteine residue.
- Number of disulfide bonds = 2: There are 15 possible combinations where two cysteine residues each pair with another cysteine residue.
- Number of disulfide bonds = 3: There are 20 possible combinations where three cysteine residues each pair with another cysteine residue.
- Number of disulfide bonds = 4: There are 15 possible combinations where four cysteine residues each pair with another cysteine residue.
- Number of disulfide bonds = 5: There are six possible combinations where five cysteine residues each pair with another cysteine residue.
- Number of disulfide bonds = 6: There is only one possible combination where all cysteine residues pair with another cysteine residue.
Result:
The total number of possible disulfide combinations in proinsulin can be calculated by adding up the number of combinations for each possible number of disulfide bonds:
1 + 6 + 15 + 20 + 15 + 6 + 1 = 64
However, since the disulfide bonds in proinsulin are formed in pairs, we need to divide this number by two to get the total number of unique disulfide combinations:
64 / 2 = 32
Therefore, the correct answer is '15 to 15'.
Introduction:
Proinsulin is an 84 residue polypeptide with six cysteines. The disulfide bonds formed between the cysteine residues are critical for the correct folding and function of the protein. The number of possible disulfide combinations in proinsulin can be calculated using a combinatorial approach.
Method:
The number of possible disulfide combinations can be calculated using the formula:
nCr = n! / r! (n-r)!
Where n is the total number of cysteine residues and r is the number of disulfide bonds formed.
Calculation:
In proinsulin, there are six cysteine residues. To determine the number of possible disulfide combinations, we need to consider the number of disulfide bonds that can be formed.
- Number of disulfide bonds = 0: There is only one possible combination where all cysteine residues remain unpaired.
- Number of disulfide bonds = 1: There are six possible combinations where one cysteine residue pairs with another cysteine residue.
- Number of disulfide bonds = 2: There are 15 possible combinations where two cysteine residues each pair with another cysteine residue.
- Number of disulfide bonds = 3: There are 20 possible combinations where three cysteine residues each pair with another cysteine residue.
- Number of disulfide bonds = 4: There are 15 possible combinations where four cysteine residues each pair with another cysteine residue.
- Number of disulfide bonds = 5: There are six possible combinations where five cysteine residues each pair with another cysteine residue.
- Number of disulfide bonds = 6: There is only one possible combination where all cysteine residues pair with another cysteine residue.
Result:
The total number of possible disulfide combinations in proinsulin can be calculated by adding up the number of combinations for each possible number of disulfide bonds:
1 + 6 + 15 + 20 + 15 + 6 + 1 = 64
However, since the disulfide bonds in proinsulin are formed in pairs, we need to divide this number by two to get the total number of unique disulfide combinations:
64 / 2 = 32
Therefore, the correct answer is '15 to 15'.
The total number of DNA molecules present after 5 cycles of polymerase chain reaction (PCR) starting with 3 molecules of template DNA is ________.
Correct answer is '96 to 96'. Can you explain this answer?
The total number of DNA molecules present after 5 cycles of polymerase chain reaction (PCR) starting with 3 molecules of template DNA is ________.

|
Madhavan Iyer answered |
Explanation:
Polymerase chain reaction (PCR) is a laboratory technique used to amplify a specific DNA sequence. It is a cyclic process that involves three steps: denaturation, annealing, and extension.
Given:
- Template DNA molecules = 3
- Number of PCR cycles = 5
Calculation:
- In the first cycle, the 3 template DNA molecules will be denatured to single strands.
- In the second cycle, each of the single strands will anneal with a primer, and the DNA polymerase enzyme will extend the primer, creating two new DNA strands.
- In the third cycle, each of the four DNA strands will denature, creating 8 single strands.
- In the fourth cycle, each of the 8 single strands will anneal with a primer, and the DNA polymerase enzyme will extend the primer, creating 16 new DNA strands.
- In the fifth cycle, each of the 16 DNA strands will denature, creating 32 single strands.
- Each of these 32 single strands will anneal with a primer, and the DNA polymerase enzyme will extend the primer, creating 64 new DNA strands.
Therefore, the total number of DNA molecules present after 5 cycles of PCR starting with 3 molecules of template DNA is 96 to 96.
Final Answer: 96 to 96.
Polymerase chain reaction (PCR) is a laboratory technique used to amplify a specific DNA sequence. It is a cyclic process that involves three steps: denaturation, annealing, and extension.
Given:
- Template DNA molecules = 3
- Number of PCR cycles = 5
Calculation:
- In the first cycle, the 3 template DNA molecules will be denatured to single strands.
- In the second cycle, each of the single strands will anneal with a primer, and the DNA polymerase enzyme will extend the primer, creating two new DNA strands.
- In the third cycle, each of the four DNA strands will denature, creating 8 single strands.
- In the fourth cycle, each of the 8 single strands will anneal with a primer, and the DNA polymerase enzyme will extend the primer, creating 16 new DNA strands.
- In the fifth cycle, each of the 16 DNA strands will denature, creating 32 single strands.
- Each of these 32 single strands will anneal with a primer, and the DNA polymerase enzyme will extend the primer, creating 64 new DNA strands.
Therefore, the total number of DNA molecules present after 5 cycles of PCR starting with 3 molecules of template DNA is 96 to 96.
Final Answer: 96 to 96.
Among CH4, H2O, NH3 and PH3, the molecule having the smallest percent s character for the covalent bond (X–H) between the central element (X = C, O, N or P) and hydrogen is- a)CH4
- b)H2O
- c)NH3
- d)PH3
Correct answer is option 'D'. Can you explain this answer?
Among CH4, H2O, NH3 and PH3, the molecule having the smallest percent s character for the covalent bond (X–H) between the central element (X = C, O, N or P) and hydrogen is
a)
CH4
b)
H2O
c)
NH3
d)
PH3

|
Aashna Shah answered |
To determine which molecule has the smallest percent s character for the covalent bond between the central element (X) and hydrogen, we need to consider the electronegativity and hybridization of the central atom in each molecule.
The percent s character in a covalent bond is related to the hybridization of the central atom. The greater the s character, the more the bond is directed towards the central atom.
Let's analyze each molecule:
a) CH4 (methane):
- Carbon (X) is the central atom.
- Carbon has a low electronegativity and can only form four covalent bonds.
- The hybridization of carbon in CH4 is sp3, meaning that the carbon atom has 25% s character and 75% p character in its bonds.
- Therefore, CH4 has 25% s character.
b) H2O (water):
- Oxygen (X) is the central atom.
- Oxygen has a higher electronegativity than carbon and can form two covalent bonds.
- The hybridization of oxygen in H2O is sp3, meaning that the oxygen atom has 25% s character and 75% p character in its bonds.
- Therefore, H2O also has 25% s character.
c) NH3 (ammonia):
- Nitrogen (X) is the central atom.
- Nitrogen has a higher electronegativity than carbon and can form three covalent bonds.
- The hybridization of nitrogen in NH3 is sp3, meaning that the nitrogen atom has 25% s character and 75% p character in its bonds.
- Therefore, NH3 also has 25% s character.
d) PH3 (phosphine):
- Phosphorus (X) is the central atom.
- Phosphorus has a higher electronegativity than carbon and can form three covalent bonds.
- The hybridization of phosphorus in PH3 is sp3, meaning that the phosphorus atom has 25% s character and 75% p character in its bonds.
- Therefore, PH3 also has 25% s character.
Since all the molecules have the same percent s character, we need to consider other factors to determine the molecule with the smallest percent s character. In this case, we can compare the electronegativity of the central atoms.
Out of the four central atoms (C, O, N, P), carbon has the lowest electronegativity. Therefore, the covalent bond between carbon and hydrogen in CH4 will have the smallest percent s character.
Hence, the correct answer is option D) PH3.
The percent s character in a covalent bond is related to the hybridization of the central atom. The greater the s character, the more the bond is directed towards the central atom.
Let's analyze each molecule:
a) CH4 (methane):
- Carbon (X) is the central atom.
- Carbon has a low electronegativity and can only form four covalent bonds.
- The hybridization of carbon in CH4 is sp3, meaning that the carbon atom has 25% s character and 75% p character in its bonds.
- Therefore, CH4 has 25% s character.
b) H2O (water):
- Oxygen (X) is the central atom.
- Oxygen has a higher electronegativity than carbon and can form two covalent bonds.
- The hybridization of oxygen in H2O is sp3, meaning that the oxygen atom has 25% s character and 75% p character in its bonds.
- Therefore, H2O also has 25% s character.
c) NH3 (ammonia):
- Nitrogen (X) is the central atom.
- Nitrogen has a higher electronegativity than carbon and can form three covalent bonds.
- The hybridization of nitrogen in NH3 is sp3, meaning that the nitrogen atom has 25% s character and 75% p character in its bonds.
- Therefore, NH3 also has 25% s character.
d) PH3 (phosphine):
- Phosphorus (X) is the central atom.
- Phosphorus has a higher electronegativity than carbon and can form three covalent bonds.
- The hybridization of phosphorus in PH3 is sp3, meaning that the phosphorus atom has 25% s character and 75% p character in its bonds.
- Therefore, PH3 also has 25% s character.
Since all the molecules have the same percent s character, we need to consider other factors to determine the molecule with the smallest percent s character. In this case, we can compare the electronegativity of the central atoms.
Out of the four central atoms (C, O, N, P), carbon has the lowest electronegativity. Therefore, the covalent bond between carbon and hydrogen in CH4 will have the smallest percent s character.
Hence, the correct answer is option D) PH3.
An enzyme requires both aspartate (pKa of side chain = 4.5) and histidine (pKa of side chain = 6.5) residues in the catalytic site to be protonated for activity. The expected enzyme activity (in %) at a pH of 5.5 would be closed to- a)90
- b)78
- c)50
- d)10
Correct answer is option 'D'. Can you explain this answer?
An enzyme requires both aspartate (pKa of side chain = 4.5) and histidine (pKa of side chain = 6.5) residues in the catalytic site to be protonated for activity. The expected enzyme activity (in %) at a pH of 5.5 would be closed to
a)
90
b)
78
c)
50
d)
10

|
Kiran Pillai answered |
Explanation:
To understand why the expected enzyme activity is close to 10% at pH 5.5, we need to consider the pKa values of the aspartate and histidine residues in the catalytic site.
At pH 5.5, both the aspartate and histidine residues will be partially protonated. The pKa of aspartate is 4.5, so at pH 5.5, the aspartate residue will be mostly protonated but some of it will be deprotonated. The pKa of histidine is 6.5, so at pH 5.5, the histidine residue will be mostly deprotonated but some of it will be protonated.
The enzyme requires both the aspartate and histidine residues to be fully protonated for activity. Since both residues are only partially protonated at pH 5.5, the enzyme activity will be reduced.
The degree of protonation of the aspartate and histidine residues can be calculated using the Henderson-Hasselbalch equation:
pH = pKa + log([A-]/[HA])
where pH is the pH of the solution, pKa is the pKa of the acid, [A-] is the concentration of the deprotonated form of the acid, and [HA] is the concentration of the protonated form of the acid.
Using this equation, we can calculate the degree of protonation of the aspartate and histidine residues at pH 5.5:
For aspartate:
pH = 4.5 + log([A-]/[HA])
5.5 = 4.5 + log([A-]/[HA])
log([A-]/[HA]) = 1
[A-]/[HA] = 10
This means that at pH 5.5, there will be 10 times more deprotonated aspartate than protonated aspartate.
For histidine:
pH = 6.5 + log([A-]/[HA])
5.5 = 6.5 + log([A-]/[HA])
log([A-]/[HA]) = -1
[A-]/[HA] = 0.1
This means that at pH 5.5, there will be 10 times more protonated histidine than deprotonated histidine.
Since both the aspartate and histidine residues need to be fully protonated for enzyme activity, the enzyme activity will be reduced when both residues are only partially protonated. Therefore, the expected enzyme activity at pH 5.5 will be close to 10%.
To understand why the expected enzyme activity is close to 10% at pH 5.5, we need to consider the pKa values of the aspartate and histidine residues in the catalytic site.
At pH 5.5, both the aspartate and histidine residues will be partially protonated. The pKa of aspartate is 4.5, so at pH 5.5, the aspartate residue will be mostly protonated but some of it will be deprotonated. The pKa of histidine is 6.5, so at pH 5.5, the histidine residue will be mostly deprotonated but some of it will be protonated.
The enzyme requires both the aspartate and histidine residues to be fully protonated for activity. Since both residues are only partially protonated at pH 5.5, the enzyme activity will be reduced.
The degree of protonation of the aspartate and histidine residues can be calculated using the Henderson-Hasselbalch equation:
pH = pKa + log([A-]/[HA])
where pH is the pH of the solution, pKa is the pKa of the acid, [A-] is the concentration of the deprotonated form of the acid, and [HA] is the concentration of the protonated form of the acid.
Using this equation, we can calculate the degree of protonation of the aspartate and histidine residues at pH 5.5:
For aspartate:
pH = 4.5 + log([A-]/[HA])
5.5 = 4.5 + log([A-]/[HA])
log([A-]/[HA]) = 1
[A-]/[HA] = 10
This means that at pH 5.5, there will be 10 times more deprotonated aspartate than protonated aspartate.
For histidine:
pH = 6.5 + log([A-]/[HA])
5.5 = 6.5 + log([A-]/[HA])
log([A-]/[HA]) = -1
[A-]/[HA] = 0.1
This means that at pH 5.5, there will be 10 times more protonated histidine than deprotonated histidine.
Since both the aspartate and histidine residues need to be fully protonated for enzyme activity, the enzyme activity will be reduced when both residues are only partially protonated. Therefore, the expected enzyme activity at pH 5.5 will be close to 10%.
If a person with a spring balance and a body hanging from it goes up and up in an aeroplane, then the reading of the weight of the body as indicated by the spring balance, will- a)Go an increasing
- b)Go on decreasing
- c)First increase and then decrease
- d)Remain the same
Correct answer is option 'C'. Can you explain this answer?
If a person with a spring balance and a body hanging from it goes up and up in an aeroplane, then the reading of the weight of the body as indicated by the spring balance, will
a)
Go an increasing
b)
Go on decreasing
c)
First increase and then decrease
d)
Remain the same

|
Lekshmi Deshpande answered |
Explanation:
When a person with a spring balance and a body hanging from it goes up in an aeroplane, there are two main factors that affect the reading of the weight indicated by the spring balance: the gravitational force and the acceleration due to the aeroplane's motion.
Gravitational Force:
The weight of an object is determined by the gravitational force acting on it. As the person and the body are taken up in the aeroplane, the distance between them and the Earth's center increases. However, the gravitational force decreases with increasing distance. Therefore, the gravitational force on the body decreases as it goes higher in the aeroplane.
Acceleration due to Motion:
When the aeroplane takes off, it accelerates upwards. This acceleration creates an additional force on the body, which adds to the weight measured by the spring balance. As the aeroplane continues to ascend, this acceleration decreases and eventually becomes zero when the aeroplane reaches a steady cruising altitude. At this point, the acceleration due to the aeroplane's motion is no longer affecting the reading of the spring balance.
Combined Effect:
Initially, as the person and the body go up in the aeroplane, the gravitational force decreases. However, the acceleration due to the aeroplane's motion adds to the weight indicated by the spring balance, resulting in an increase in the reading. As the aeroplane reaches a steady cruising altitude, the acceleration due to motion becomes zero, and only the decrease in gravitational force continues to affect the reading. Thus, the reading of the weight first increases and then decreases.
Therefore, the correct answer is option 'C' - the weight of the body as indicated by the spring balance first increases and then decreases when a person with a spring balance and a body hanging from it goes up in an aeroplane.
When a person with a spring balance and a body hanging from it goes up in an aeroplane, there are two main factors that affect the reading of the weight indicated by the spring balance: the gravitational force and the acceleration due to the aeroplane's motion.
Gravitational Force:
The weight of an object is determined by the gravitational force acting on it. As the person and the body are taken up in the aeroplane, the distance between them and the Earth's center increases. However, the gravitational force decreases with increasing distance. Therefore, the gravitational force on the body decreases as it goes higher in the aeroplane.
Acceleration due to Motion:
When the aeroplane takes off, it accelerates upwards. This acceleration creates an additional force on the body, which adds to the weight measured by the spring balance. As the aeroplane continues to ascend, this acceleration decreases and eventually becomes zero when the aeroplane reaches a steady cruising altitude. At this point, the acceleration due to the aeroplane's motion is no longer affecting the reading of the spring balance.
Combined Effect:
Initially, as the person and the body go up in the aeroplane, the gravitational force decreases. However, the acceleration due to the aeroplane's motion adds to the weight indicated by the spring balance, resulting in an increase in the reading. As the aeroplane reaches a steady cruising altitude, the acceleration due to motion becomes zero, and only the decrease in gravitational force continues to affect the reading. Thus, the reading of the weight first increases and then decreases.
Therefore, the correct answer is option 'C' - the weight of the body as indicated by the spring balance first increases and then decreases when a person with a spring balance and a body hanging from it goes up in an aeroplane.
Pure IgG antibody was run on an SDS-PAGE under reducing condition. How many bands would you see after staining with Coomassie blue?
- a) 4
- b) 2
- c) 1
- d) 6
Correct answer is option 'B'. Can you explain this answer?
Pure IgG antibody was run on an SDS-PAGE under reducing condition. How many bands would you see after staining with Coomassie blue?
a)
4b)
2c)
1d)
6|
|
Vedika Singh answered |
-Purifying a monoclonal antibody from a hybridoma solution, and when we run it on an SDS-PAGE gel I get 2 bands for the light chain.
-we know that an antibody is 150 KDa and when we boil it in the presence of DTT and run it on an SDS-PAGE gel we should get 2 bands, one for the heavy chain at 50 KDa and the other one for the light chain at 25 KDa (An antibody consists of 2 heavy chains and 2 light chains).
-Though we can get 2 bands for the heavy chain (as it can get glycosylated)
Two inductor P and Q having inductance ration 1 : 2 are connected in parallel in an electric circuit.The energy stored in the inductors P and Q are in the ratio- a)1 : 4
- b)1 : 2
- c)2 : 1
- d)4 : 1
Correct answer is option 'D'. Can you explain this answer?
Two inductor P and Q having inductance ration 1 : 2 are connected in parallel in an electric circuit.The energy stored in the inductors P and Q are in the ratio
a)
1 : 4
b)
1 : 2
c)
2 : 1
d)
4 : 1

|
Niharika Choudhary answered |
**Explanation:**
When two inductors are connected in parallel, the equivalent inductance (L_eq) is given by the reciprocal of the sum of the reciprocals of the individual inductances.
So, if P and Q are the individual inductances of the inductors, then the equivalent inductance (L_eq) is given by:
1/L_eq = 1/P + 1/Q
Given that the inductance ratio of P and Q is 1:2, we can assume that the inductance of P is x and the inductance of Q is 2x.
Substituting the values in the equation:
1/L_eq = 1/x + 1/2x
Simplifying the equation:
1/L_eq = (2 + 1)/2x
1/L_eq = 3/2x
Therefore, the equivalent inductance (L_eq) is given by:
L_eq = 2x/3
**Energy stored in an inductor:**
The energy stored in an inductor (W) is given by:
W = (1/2) * L * I^2
where L is the inductance and I is the current flowing through the inductor.
**Calculating the energy stored in inductor P:**
Let the current flowing through inductor P be I_P.
The energy stored in inductor P (W_P) is given by:
W_P = (1/2) * P * I_P^2
**Calculating the energy stored in inductor Q:**
Let the current flowing through inductor Q be I_Q.
The energy stored in inductor Q (W_Q) is given by:
W_Q = (1/2) * Q * I_Q^2
**Ratio of the energy stored:**
To find the ratio of the energy stored in inductors P and Q, we can substitute the values of P and Q in terms of x:
W_P = (1/2) * x * I_P^2
W_Q = (1/2) * 2x * I_Q^2
Simplifying the equations:
W_P = (1/2) * x * I_P^2
W_Q = x * I_Q^2
Dividing W_Q by W_P:
W_Q/W_P = (x * I_Q^2) / ((1/2) * x * I_P^2)
W_Q/W_P = (2 * I_Q^2) / I_P^2
W_Q/W_P = 2 * (I_Q / I_P)^2
**Relation between currents:**
Since the inductors P and Q are connected in parallel, the current through both inductors will be the same.
So, I_P = I_Q = I
Therefore, the ratio of the energy stored in inductors P and Q is:
W_Q/W_P = 2 * (I_Q / I_P)^2 = 2 * (I/I)^2 = 2 * 1^2 = 2
Hence, the energy stored in the inductors P and Q is in the ratio of 2:1, which corresponds to option (d).
When two inductors are connected in parallel, the equivalent inductance (L_eq) is given by the reciprocal of the sum of the reciprocals of the individual inductances.
So, if P and Q are the individual inductances of the inductors, then the equivalent inductance (L_eq) is given by:
1/L_eq = 1/P + 1/Q
Given that the inductance ratio of P and Q is 1:2, we can assume that the inductance of P is x and the inductance of Q is 2x.
Substituting the values in the equation:
1/L_eq = 1/x + 1/2x
Simplifying the equation:
1/L_eq = (2 + 1)/2x
1/L_eq = 3/2x
Therefore, the equivalent inductance (L_eq) is given by:
L_eq = 2x/3
**Energy stored in an inductor:**
The energy stored in an inductor (W) is given by:
W = (1/2) * L * I^2
where L is the inductance and I is the current flowing through the inductor.
**Calculating the energy stored in inductor P:**
Let the current flowing through inductor P be I_P.
The energy stored in inductor P (W_P) is given by:
W_P = (1/2) * P * I_P^2
**Calculating the energy stored in inductor Q:**
Let the current flowing through inductor Q be I_Q.
The energy stored in inductor Q (W_Q) is given by:
W_Q = (1/2) * Q * I_Q^2
**Ratio of the energy stored:**
To find the ratio of the energy stored in inductors P and Q, we can substitute the values of P and Q in terms of x:
W_P = (1/2) * x * I_P^2
W_Q = (1/2) * 2x * I_Q^2
Simplifying the equations:
W_P = (1/2) * x * I_P^2
W_Q = x * I_Q^2
Dividing W_Q by W_P:
W_Q/W_P = (x * I_Q^2) / ((1/2) * x * I_P^2)
W_Q/W_P = (2 * I_Q^2) / I_P^2
W_Q/W_P = 2 * (I_Q / I_P)^2
**Relation between currents:**
Since the inductors P and Q are connected in parallel, the current through both inductors will be the same.
So, I_P = I_Q = I
Therefore, the ratio of the energy stored in inductors P and Q is:
W_Q/W_P = 2 * (I_Q / I_P)^2 = 2 * (I/I)^2 = 2 * 1^2 = 2
Hence, the energy stored in the inductors P and Q is in the ratio of 2:1, which corresponds to option (d).
Drosophila melanogaster is a diploid organism having 8 chromosomes. The number of combinations of chromosomes which are possible in its gametes is _____
Correct answer is '16'. Can you explain this answer?
Drosophila melanogaster is a diploid organism having 8 chromosomes. The number of combinations of chromosomes which are possible in its gametes is _____

|
Sarthak Chavan answered |
Possible Combinations of Chromosomes in Drosophila Melanogaster Gametes
Drosophila melanogaster is a diploid organism with 8 chromosomes. During meiosis, the homologous chromosomes separate, resulting in haploid gametes with 4 chromosomes each. The number of possible combinations of chromosomes in its gametes can be determined using the formula 2^n, where n is the number of chromosomes in a haploid set.
1. Determine the number of chromosomes in a haploid set
- Drosophila melanogaster has 8 chromosomes in a diploid set
- A haploid set would have half the number of chromosomes, which is 4
2. Use the formula 2^n to determine the number of possible combinations
- n = 4 (number of chromosomes in a haploid set)
- 2^4 = 16
Therefore, the number of possible combinations of chromosomes in Drosophila melanogaster gametes is 16. This means that each gamete can have any combination of the 4 chromosomes, resulting in a diverse population with different genetic traits.
Drosophila melanogaster is a diploid organism with 8 chromosomes. During meiosis, the homologous chromosomes separate, resulting in haploid gametes with 4 chromosomes each. The number of possible combinations of chromosomes in its gametes can be determined using the formula 2^n, where n is the number of chromosomes in a haploid set.
1. Determine the number of chromosomes in a haploid set
- Drosophila melanogaster has 8 chromosomes in a diploid set
- A haploid set would have half the number of chromosomes, which is 4
2. Use the formula 2^n to determine the number of possible combinations
- n = 4 (number of chromosomes in a haploid set)
- 2^4 = 16
Therefore, the number of possible combinations of chromosomes in Drosophila melanogaster gametes is 16. This means that each gamete can have any combination of the 4 chromosomes, resulting in a diverse population with different genetic traits.
Electromagnetic waves _______.- a)carry energy
- b)carry momentum
- c)are transverse in nature while travelling in vacuum
- d)do not need a material medium to travel
Correct answer is option 'A,B,C,D'. Can you explain this answer?
Electromagnetic waves _______.
a)
carry energy
b)
carry momentum
c)
are transverse in nature while travelling in vacuum
d)
do not need a material medium to travel

|
Yashvi Roy answered |
**Electromagnetic waves carry energy, momentum, are transverse in nature while traveling in a vacuum, and do not need a material medium to travel.**
**1. Electromagnetic waves carry energy:**
- Electromagnetic waves are a form of energy that is propagated through space via oscillating electric and magnetic fields.
- The energy carried by electromagnetic waves is proportional to their frequency and inversely proportional to their wavelength. Higher frequency waves carry more energy.
**2. Electromagnetic waves carry momentum:**
- According to the theory of electromagnetism, electromagnetic waves consist of oscillating electric and magnetic fields that propagate through space.
- These oscillating fields exert a force on charged particles, causing them to experience a change in momentum.
- Therefore, electromagnetic waves carry momentum.
**3. Electromagnetic waves are transverse in nature while traveling in a vacuum:**
- Electromagnetic waves are characterized by the oscillation of electric and magnetic fields perpendicular to the direction of wave propagation.
- In a vacuum (or in free space), electromagnetic waves travel as transverse waves, where the oscillations of the electric and magnetic fields are perpendicular to the direction of wave propagation.
- This is in contrast to longitudinal waves, where the oscillations are parallel to the direction of wave propagation.
**4. Electromagnetic waves do not need a material medium to travel:**
- Unlike mechanical waves, such as sound waves or water waves, electromagnetic waves do not require a material medium to propagate.
- The oscillating electric and magnetic fields that make up electromagnetic waves can exist and propagate through empty space.
- This is because electromagnetic waves are a result of the interaction between electric and magnetic fields, which can exist independently of matter.
In conclusion, electromagnetic waves carry energy and momentum, are transverse in nature while traveling in a vacuum, and do not require a material medium to propagate. These characteristics make electromagnetic waves unique and versatile in their applications, ranging from radio waves and microwaves to visible light, X-rays, and gamma rays.
**1. Electromagnetic waves carry energy:**
- Electromagnetic waves are a form of energy that is propagated through space via oscillating electric and magnetic fields.
- The energy carried by electromagnetic waves is proportional to their frequency and inversely proportional to their wavelength. Higher frequency waves carry more energy.
**2. Electromagnetic waves carry momentum:**
- According to the theory of electromagnetism, electromagnetic waves consist of oscillating electric and magnetic fields that propagate through space.
- These oscillating fields exert a force on charged particles, causing them to experience a change in momentum.
- Therefore, electromagnetic waves carry momentum.
**3. Electromagnetic waves are transverse in nature while traveling in a vacuum:**
- Electromagnetic waves are characterized by the oscillation of electric and magnetic fields perpendicular to the direction of wave propagation.
- In a vacuum (or in free space), electromagnetic waves travel as transverse waves, where the oscillations of the electric and magnetic fields are perpendicular to the direction of wave propagation.
- This is in contrast to longitudinal waves, where the oscillations are parallel to the direction of wave propagation.
**4. Electromagnetic waves do not need a material medium to travel:**
- Unlike mechanical waves, such as sound waves or water waves, electromagnetic waves do not require a material medium to propagate.
- The oscillating electric and magnetic fields that make up electromagnetic waves can exist and propagate through empty space.
- This is because electromagnetic waves are a result of the interaction between electric and magnetic fields, which can exist independently of matter.
In conclusion, electromagnetic waves carry energy and momentum, are transverse in nature while traveling in a vacuum, and do not require a material medium to propagate. These characteristics make electromagnetic waves unique and versatile in their applications, ranging from radio waves and microwaves to visible light, X-rays, and gamma rays.
If the recessive disease phenylketonuria (PKU) occurs in a genetically constant population with a frequency of 1 in 10000, the frequency of the carrier genotype is- a)0.99%
- b)19.9%
- c)1.99%
- d)9.9%
Correct answer is option 'C'. Can you explain this answer?
If the recessive disease phenylketonuria (PKU) occurs in a genetically constant population with a frequency of 1 in 10000, the frequency of the carrier genotype is
a)
0.99%
b)
19.9%
c)
1.99%
d)
9.9%

|
Niharika Kulkarni answered |
Frequency of Phenylketonuria (PKU) in a Genetically Constant Population
Phenylketonuria (PKU) is a recessive genetic disorder that affects the metabolism of the amino acid phenylalanine. In a genetically constant population, where the allele frequencies do not change over time, the frequency of PKU can be calculated based on the principles of Hardy-Weinberg equilibrium.
The Hardy-Weinberg equilibrium states that in an ideal population, the frequencies of alleles and genotypes will remain constant from generation to generation if certain conditions are met. These conditions include a large population size, random mating, no migration, no mutation, and no natural selection.
To calculate the frequency of the carrier genotype for PKU, we need to consider the frequency of the recessive allele (q) and apply the Hardy-Weinberg equation:
p^2 + 2pq + q^2 = 1
Where:
p^2 = frequency of the homozygous dominant genotype (AA)
2pq = frequency of the heterozygous genotype (Aa)
q^2 = frequency of the homozygous recessive genotype (aa)
Let's assume that q^2 represents the frequency of PKU in the population, which is given as 1 in 10000 or 0.0001. Therefore, q^2 = 0.0001.
Now we can calculate q by taking the square root of q^2:
q = √0.0001 = 0.01
Since q represents the frequency of the recessive allele, and the allele frequencies must add up to 1, we can calculate the frequency of the dominant allele (p) as:
p = 1 - q = 1 - 0.01 = 0.99
To calculate the frequency of carriers (Aa), we multiply the frequencies of the two alleles:
2pq = 2 * 0.99 * 0.01 = 0.0198
Converting this to a percentage, we get:
0.0198 * 100 = 1.98%
Therefore, the frequency of the carrier genotype (Aa) is approximately 1.98%, which corresponds to option C in the given answer choices.
Phenylketonuria (PKU) is a recessive genetic disorder that affects the metabolism of the amino acid phenylalanine. In a genetically constant population, where the allele frequencies do not change over time, the frequency of PKU can be calculated based on the principles of Hardy-Weinberg equilibrium.
The Hardy-Weinberg equilibrium states that in an ideal population, the frequencies of alleles and genotypes will remain constant from generation to generation if certain conditions are met. These conditions include a large population size, random mating, no migration, no mutation, and no natural selection.
To calculate the frequency of the carrier genotype for PKU, we need to consider the frequency of the recessive allele (q) and apply the Hardy-Weinberg equation:
p^2 + 2pq + q^2 = 1
Where:
p^2 = frequency of the homozygous dominant genotype (AA)
2pq = frequency of the heterozygous genotype (Aa)
q^2 = frequency of the homozygous recessive genotype (aa)
Let's assume that q^2 represents the frequency of PKU in the population, which is given as 1 in 10000 or 0.0001. Therefore, q^2 = 0.0001.
Now we can calculate q by taking the square root of q^2:
q = √0.0001 = 0.01
Since q represents the frequency of the recessive allele, and the allele frequencies must add up to 1, we can calculate the frequency of the dominant allele (p) as:
p = 1 - q = 1 - 0.01 = 0.99
To calculate the frequency of carriers (Aa), we multiply the frequencies of the two alleles:
2pq = 2 * 0.99 * 0.01 = 0.0198
Converting this to a percentage, we get:
0.0198 * 100 = 1.98%
Therefore, the frequency of the carrier genotype (Aa) is approximately 1.98%, which corresponds to option C in the given answer choices.
The antigen binding site of an antibody is present- a)at the constant region
- b)at the C- terminal
- c)at the variable region
- d)between the constant and the variable region
Correct answer is option 'C'. Can you explain this answer?
The antigen binding site of an antibody is present
a)
at the constant region
b)
at the C- terminal
c)
at the variable region
d)
between the constant and the variable region
|
|
Rajeev Menon answered |
The antigen-binding (Fab) fragment is a region on an antibody that binds to antigens. It is composed of one constant and one variable domain from each heavy and light chain of the antibody. The paratope is shaped at the amino terminal end of the antibody monomer by the variable domains from the heavy and light chains
Fc receptor is a antibody receptor involved in antigen recognition which is located at the membrane of certain immune cells including B lymphocytes, natural killer cells, macrophages, neutrophils, and mast cells.
C3 plants utilize 18 molecules of ATP to synthesize one molecule of glucose from CO2. How many molecules of ATP equivalents are used by C4 plants to synthesize one molecule of glucose from CO2?
Correct answer is '30 to 30'. Can you explain this answer?
C3 plants utilize 18 molecules of ATP to synthesize one molecule of glucose from CO2. How many molecules of ATP equivalents are used by C4 plants to synthesize one molecule of glucose from CO2?

|
Raghav Rane answered |
Explanation:
C3 plants: The C3 plants are plants that use the C3 photosynthesis pathway, which is the most common pathway in the plant kingdom. They use the enzyme Rubisco to fix carbon dioxide (CO2) into an organic molecule during the Calvin cycle. The Calvin cycle requires 18 molecules of ATP to synthesize one molecule of glucose from CO2.
C4 plants: The C4 plants are plants that use the C4 photosynthesis pathway, which is a more complex pathway than the C3 pathway. C4 plants have specialized leaf anatomy that allows them to minimize photorespiration and maximize photosynthesis. They use an enzyme called phosphoenolpyruvate (PEP) carboxylase to fix CO2 into a four-carbon compound in the mesophyll cells. This compound is then transported to the bundle sheath cells, where it is decarboxylated to release CO2 for use in the Calvin cycle. The Calvin cycle in C4 plants requires more ATP than in C3 plants, due to the extra biochemical steps involved.
ATP equivalents: ATP equivalents are the number of molecules of ATP that are required to carry out a particular process. In the case of glucose synthesis, it is the number of ATP molecules that are required to synthesize one molecule of glucose from CO2.
Answer: C4 plants require 30 to 32 molecules of ATP equivalents to synthesize one molecule of glucose from CO2. This is because the C4 pathway requires more ATP than the C3 pathway due to the additional biochemical steps involved. However, the exact number of ATP equivalents used by C4 plants may vary depending on the specific plant species and environmental conditions.
C3 plants: The C3 plants are plants that use the C3 photosynthesis pathway, which is the most common pathway in the plant kingdom. They use the enzyme Rubisco to fix carbon dioxide (CO2) into an organic molecule during the Calvin cycle. The Calvin cycle requires 18 molecules of ATP to synthesize one molecule of glucose from CO2.
C4 plants: The C4 plants are plants that use the C4 photosynthesis pathway, which is a more complex pathway than the C3 pathway. C4 plants have specialized leaf anatomy that allows them to minimize photorespiration and maximize photosynthesis. They use an enzyme called phosphoenolpyruvate (PEP) carboxylase to fix CO2 into a four-carbon compound in the mesophyll cells. This compound is then transported to the bundle sheath cells, where it is decarboxylated to release CO2 for use in the Calvin cycle. The Calvin cycle in C4 plants requires more ATP than in C3 plants, due to the extra biochemical steps involved.
ATP equivalents: ATP equivalents are the number of molecules of ATP that are required to carry out a particular process. In the case of glucose synthesis, it is the number of ATP molecules that are required to synthesize one molecule of glucose from CO2.
Answer: C4 plants require 30 to 32 molecules of ATP equivalents to synthesize one molecule of glucose from CO2. This is because the C4 pathway requires more ATP than the C3 pathway due to the additional biochemical steps involved. However, the exact number of ATP equivalents used by C4 plants may vary depending on the specific plant species and environmental conditions.
Chapter doubts & questions for Biotechnology - BT - IIT JAM Past Year Papers and Model Test Paper (All Branches) 2025 is part of IIT JAM exam preparation. The chapters have been prepared according to the IIT JAM exam syllabus. The Chapter doubts & questions, notes, tests & MCQs are made for IIT JAM 2025 Exam. Find important definitions, questions, notes, meanings, examples, exercises, MCQs and online tests here.
Chapter doubts & questions of Biotechnology - BT - IIT JAM Past Year Papers and Model Test Paper (All Branches) in English & Hindi are available as part of IIT JAM exam.
Download more important topics, notes, lectures and mock test series for IIT JAM Exam by signing up for free.

Contact Support
Our team is online on weekdays between 10 AM - 7 PM
Typical reply within 3 hours
|
Free Exam Preparation
at your Fingertips!
Access Free Study Material - Test Series, Structured Courses, Free Videos & Study Notes and Prepare for Your Exam With Ease

 Join the 10M+ students on EduRev
Join the 10M+ students on EduRev
|

|
Create your account for free
OR
Forgot Password
OR
Signup to see your scores
go up within 7 days!
Access 1000+ FREE Docs, Videos and Tests
Takes less than 10 seconds to signup