NEET Exam > NEET Questions > Does chargaff's rule is applicable for D s RN...
Start Learning for Free
Does chargaff's rule is applicable for D s RNA?
Verified Answer
Does chargaff's rule is applicable for D s RNA?
It largely would, depending on your definition of ‘double stranded’. If the great majority of the RNA formed an un-gapped, continuous double helix, then it would be MOSTLY GC AU pairs, so the rule would apply.
But RNA helices are tolerant of GU pairs… so the amount of GU pairing would throw the GC AU ratios off
If we’re not talking GENOMES, but ‘any functional double-stranded RNA’ then tRNA would’ve given Chargaff nightmares! Look at this image
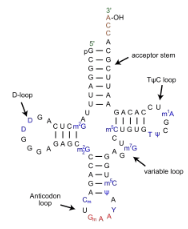
Note FIRST that there are large bits (the ‘loops’ primarily) that are NOT double-stranded, so these can throw off the ratios bc they would be ‘un-partnered’
SECOND, note that tRNAs contain a LOT of modified bases (non A, G, C, U). These are synthesized as AGCU, but modified AFTER synthesis to be weirdos. So Chargaff would’ve had more than 4 ‘players’ if he was studying tRNA.
Chargaff’s rule is a straightforward consequence of two things:
G pairs with C, A pairs with T EXCLUSIVELY in DNA
Genomic DNA is essentially exclusively double stranded, so there would be no regions with exceptions in it (not looking at you, telomeres)
 This question is part of UPSC exam. View all NEET courses
This question is part of UPSC exam. View all NEET courses
Most Upvoted Answer
Does chargaff's rule is applicable for D s RNA?
Noo
Attention NEET Students!
To make sure you are not studying endlessly, EduRev has designed NEET study material, with Structured Courses, Videos, & Test Series. Plus get personalized analysis, doubt solving and improvement plans to achieve a great score in NEET.

|
Explore Courses for NEET exam
|

|
Does chargaff's rule is applicable for D s RNA?
Question Description
Does chargaff's rule is applicable for D s RNA? for NEET 2024 is part of NEET preparation. The Question and answers have been prepared according to the NEET exam syllabus. Information about Does chargaff's rule is applicable for D s RNA? covers all topics & solutions for NEET 2024 Exam. Find important definitions, questions, meanings, examples, exercises and tests below for Does chargaff's rule is applicable for D s RNA?.
Does chargaff's rule is applicable for D s RNA? for NEET 2024 is part of NEET preparation. The Question and answers have been prepared according to the NEET exam syllabus. Information about Does chargaff's rule is applicable for D s RNA? covers all topics & solutions for NEET 2024 Exam. Find important definitions, questions, meanings, examples, exercises and tests below for Does chargaff's rule is applicable for D s RNA?.
Solutions for Does chargaff's rule is applicable for D s RNA? in English & in Hindi are available as part of our courses for NEET.
Download more important topics, notes, lectures and mock test series for NEET Exam by signing up for free.
Here you can find the meaning of Does chargaff's rule is applicable for D s RNA? defined & explained in the simplest way possible. Besides giving the explanation of
Does chargaff's rule is applicable for D s RNA?, a detailed solution for Does chargaff's rule is applicable for D s RNA? has been provided alongside types of Does chargaff's rule is applicable for D s RNA? theory, EduRev gives you an
ample number of questions to practice Does chargaff's rule is applicable for D s RNA? tests, examples and also practice NEET tests.

|
Explore Courses for NEET exam
|

|
Suggested Free Tests
Signup for Free!
Signup to see your scores go up within 7 days! Learn & Practice with 1000+ FREE Notes, Videos & Tests.























